Significance
The NLRP3 inflammasome is an innate sensor activated by signals released from pathogens or injured tissues. Activation of the NLRP3 inflammasome can be beneficial during infection and vaccination. Nonetheless, when NLRP3 activity is uncontrolled and chronic it becomes detrimental and contributes to inflammation-driven pathology in several diseases. Licensing mechanisms must exist that prevent unwanted NLRP3 inflammasome responses. Here, we characterize one such mechanism. We describe that TBK1 and IKKε, two closely related kinases activated upon TLR signaling, act as a novel OFF switch for the NLRP3 pathway. Using pharmacological and genetic approaches, we show that TBK1 and IKKε together limit the responses downstream of the NLRP3 inflammasome activation and work against the PP2A phosphatase ON switch to balance NLRP3 activity.
Keywords: NLRP3 inflammasome, macrophages, TBK1, IKKε, PP2A
Abstract
NACHT, LRR, and PYD domains–containing protein 3 (NLRP3) inflammasome activation is beneficial during infection and vaccination but, when uncontrolled, is detrimental and contributes to inflammation-driven pathologies. Hence, discovering endogenous mechanisms that regulate NLRP3 activation is important for disease interventions. Activation of NLRP3 is regulated at the transcriptional level and by posttranslational modifications. Here, we describe a posttranslational phospho-switch that licenses NLRP3 activation in macrophages. The ON switch is controlled by the protein phosphatase 2A (PP2A) downstream of a variety of NLRP3 activators in vitro and in lipopolysaccharide-induced peritonitis in vivo. The OFF switch is regulated by two closely related kinases, TANK-binding kinase 1 (TBK1) and I-kappa-B kinase epsilon (IKKε). Pharmacological inhibition of TBK1 and IKKε, as well as simultaneous deletion of TBK1 and IKKε, but not of either kinase alone, increases NLRP3 activation. In addition, TBK1/IKKε inhibitors counteract the effects of PP2A inhibition on inflammasome activity. We find that, mechanistically, TBK1 interacts with NLRP3 and controls the pathway activity at a site distinct from NLRP3-serine 3, previously reported to be under PP2A control. Mutagenesis of NLRP3 confirms serine 3 as an important phospho-switch site but, surprisingly, reveals that this is not the sole site regulated by either TBK1/IKKε or PP2A, because all retain the control over the NLRP3 pathway even when serine 3 is mutated. Altogether, a model emerges whereby TLR-activated TBK1 and IKKε act like a “parking brake” for NLRP3 activation at the time of priming, while PP2A helps remove this parking brake in the presence of NLRP3 activating signals, such as bacterial pore-forming toxins or endogenous danger signals.
NACHT, LRR, and PYD domains–containing protein 3 (NLRP3) inflammasome is a cytosolic innate sensor that detects the loss of cellular homeostasis induced by pathogens or tissue injury (1). Activation of innate sensors, including NLRP3, is critical for innate instruction of adaptive immunity (2). The NLRP3 inflammasome function is thus important for antimicrobial defenses and for the success of vaccine adjuvants, particularly those that contain a mixture of synthetic TLR4 agonists and NLRP3 activators (e.g., GLA-SE and MPL-QS21) (3–7). NLRP3 activators are structurally diverse and how they are sensed remains poorly understood. However, they all promote oligomerization of NLRP3 and its association with the effector enzyme, procaspase-1, through the adaptor molecule ASC (apoptosis-associated speck-like protein containing a CARD) (8). Clustering of procaspase-1 within the inflammasome complex results in its activation, self-cleavage, and the cleavage and secretion of key proinflammatory cytokines such as interleukin (IL)-1β and IL-18. In many cases, a cell with activated inflammasome dies through caspase-1–directed osmotic lysis (“pyroptosis”) and, thereby, releases cytoplasmic proteins that serve as alarmins (9). This initiates an inflammatory response that recruits more phagocytes to the site of insult to destroy invading pathogens and to remove dead cells and debris (1). If the inciting agent is cleared, the NLRP3 response wanes and the homeostatic state is restored.
NLRP3 activity is generally transient and beneficial in antimicrobial defenses and vaccine responses. By contrast, in inherited or acquired inflammatory conditions, particularly those associated with aging (e.g., atherosclerosis, Parkinson’s disease, and Alzheimer’s disease), protracted NLRP3 activation leads to or contributes to a pathological state (1, 10–13). In inherited diseases, NLRP3 activity is triggered by NLRP3-activating mutations (14). In acquired inflammatory diseases, NLRP3 is inadvertently triggered by the loss of cell homeostasis, for example due to the accumulation of reactive oxygen species, or by membrane damage in myeloid cells during phagocytosis of protein aggregates such as organic crystals (e.g., cholesterol and uric acid), α-synuclein, or fibrillar amyloid-β (8). In many conditions, myeloid cells have been identified as drivers of inflammation and the deletion of NLRP3 has been shown to be protective in preclinical models (10–12).
The current clinical treatment strategy to inhibit the NLRP3 pathway is to block the downstream IL-1β signaling using IL-1 receptor antagonists or IL-1β antibodies (15). This strategy has not always been effective, most likely because it targets only one of the several mediators of the inflammasome activation response. Logically, then, the target to fully control NLRP3 inflammasome activation selectively would be NLRP3 itself.
Activation of NLRP3 is regulated transcriptionally, to control messenger RNA (mRNA) expression (16, 17), posttranscriptionally, to control mRNA stability (18), and by several posttranslational modifications (PTMs), to control protein activity (8, 19). The latter are critical especially in chronic conditions where persistent NLRP3 expression occurs and NLRP3 transcriptional control is lost (20–22). PTMs, such as ubiquitination (23–29), phosphorylation (30–35), SUMOylation (36), and acetylation (37), prime NLRP3 for activation, while also maintaining it in a signaling-incompetent state in the absence of an activating signal (8). For example, NLRP3 deubiquitination in response to priming and activating signals permits inflammasome assembly (23, 24, 28). Recently, it was reported that in immortalized bone marrow–derived macrophages (iBMDMs) phosphorylation of serine 5 (S5 in human, S3 in mouse) blocks the association of NLRP3 with ASC and, thereby, inhibits the formation of the inflammasome (32). Conversely, upon inflammasome activation, NLRP3-S5 is dephosphorylated by the phosphatase PP2A (protein phosphatase 2A) to license NLRP3 activation (32).
The report by Stutz et al. (32) offered a new strategy to control NLRP3 activation but raised several important questions: How conserved is the ON switch, does it work in vivo, and which kinase controls the OFF switch? In addressing these questions, we describe here the in vivo relevance of the PP2A ON switch and its ability to control NLRP3 activation downstream of most known NLRP3 priming and activating signals. We also report that the two closely related kinases TBK1 (TANK-binding kinase 1) and IKKε (I-kappa-B kinase epsilon) act like an OFF switch for the NLRP3 pathway that counteracts the PP2A ON switch phosphatase to balance NLRP3 activity. Importantly, TBK1 and IKKε activation is induced downstream of TLRs that prime NLRP3 expression, suggesting that the same signals/kinases that prime NLRP3 expression also introduce brakes to prevent its premature activation without a bona fide signal 2. The serine/threonine kinase AKT was recently reported to inhibit NLRP3 through phosphorylation on NLRP3-serine 5 in humans (serine 3 in mice) such that NLRP3 activity is attenuated in metabolically active cells (38). Mechanistically, we find that TBK1 and IKKε control the NLRP3 pathway, in part by controlling the activity of AKT, but also that TBK1/IKKε control extends beyond AKT. First, TBK1 interacts with NLRP3 even when AKT is inhibited and when the AKT-target site, NLRP3-serine 3, is mutated. Second, our mutagenesis of NLRP3 confirms serine 3 as an important phospho-switch site, but it surprisingly reveals that, unlike for AKT (38), this is not the sole site in the NLRP3 complex regulated by either TBK1/IKKε or PP2A, as all retain control over the NLRP3 pathway activity even when serine 3 is mutated. The discovery of endogenous phospho-switches that control NLRP3 activation offers potential drug targeting opportunities to control dysregulated NLRP3-driven conditions.
Results
PP2A Phosphatase ON Switch Licenses the NLRP3 Inflammasome Activity Downstream of Multiple TLRs in Mouse and Human Macrophages.
PP2A’s modulation of NLRP3 activity was first described using iBMDMs (32). It remained unclear how conserved PP2A’s control is among a variety of inflammasome stimuli, other inflammasomes, in vivo and between species. To address these questions, we induced inflammasome activation in mouse primary BMDMs or human blood monocyte–derived macrophages (HMDMs). We primed BMDMs to induce NLRP3 and pro–IL-1β expression using several TLR ligands serving as the inflammasome-priming signal 1: lipopolysaccharide (LPS) (TLR4 ligand), zymosan (TLR2/dectin-1 ligand), or Pam3CSK4 (TLR2/TLR1 ligand) for 4 h or Poly(I:C) (TLR3 ligand) for 1 h. We induced the activation of the NLRP3 inflammasome using either the K+ efflux–dependent signals 2, the bacterial pore-forming toxin nigericin for 1 h (39) or the endogenous particle monosodium urate (MSU) for 5 h (40), or using the K+ efflux-independent signal 2, TLR7 agonist R837 for 3 h (41) (Fig. 1A). The PP2A inhibitor okadaic acid (32) was added 15 to 30 min before signal 2 (Fig. 1A). The specificity of okadaic acid for the PP2A phosphatase has been previously described as ∼100- to 200-fold higher vs. PP1 (42). A role of PP2A in NLRP3 activation downstream of LPS and nigericin activation has also been validated in immortalized macrophages using PP2A small interfering RNA (siRNA) (32).
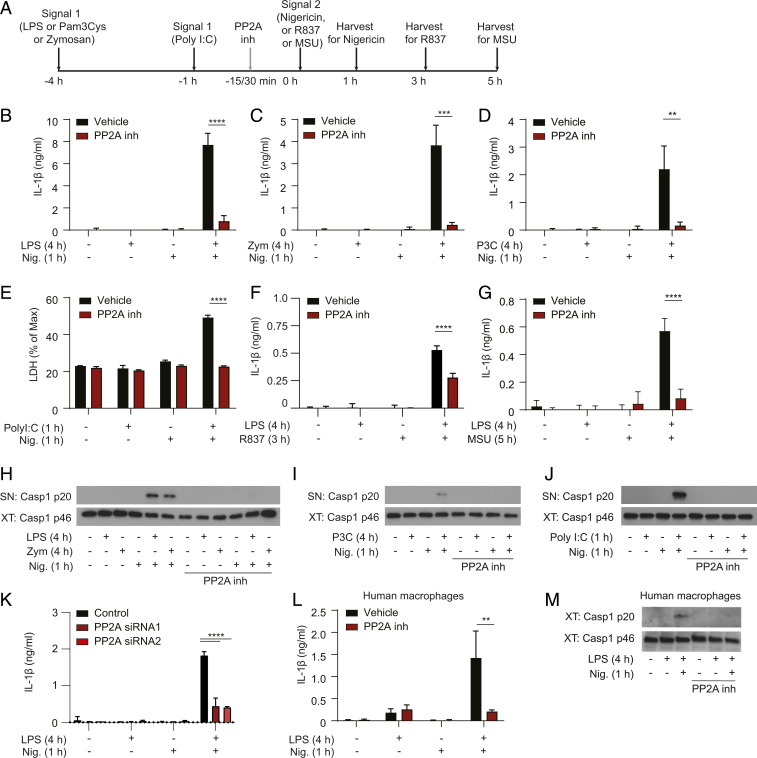
Fig. 1.
PP2A acts as a human/mouse conserved ON switch to license NLRP3 responses downstream of multiple TLRs. (A) BMDMs were primed with 100 ng/mL ultrapure LPS, 1 µg/mL Pam3CSK4, and 100 µg/mL zymosan for 4 h or 2.5 µg/mL Poly(I:C) for 1 h, followed by stimulation with 5 µM nigericin for 1 h or 70 µM R837 for 3 h or 150 µg/mL MSU for 5 h. Cells were treated with the PP2A inhibitor okadaic acid (1 µM) for 15 to 30 min before nigericin, R837, or MSU stimulation. After stimulation, cell supernatants were collected and cells lysed. (B–D, F, and G) IL-1β cytokine secretion was measured in the BMDM supernatants using ELISA. Because IL-1β is not transcriptionally induced after 1 h of Poly(I:C) priming [optimal priming time for Poly(I:C)], inflammasome-dependent cell death was used as a readout instead, measured by LDH release assay in E. (H–J) Caspase-1 was detected in BMDM supernatants (SN) and cell lysates (XT) using Western blot. One representative Western blot is shown. IL-1β data are shown as mean + SEM from pooled results of three independent experiments, except in B where data are shown as mean + SEM from two independent experiments and E where LDH data are mean + SD of triplicates from one experiment and in F where IL-1β data are mean + SD of triplicates from one experiment representative of four. (K) IL-1β cytokine secretion after PP2A knockdown was measured in iBMDM supernatants using ELISA. Data are shown as a mean + SD of triplicates from one representative of three independent experiments. In conditions where not enough cells were available for triplicates, duplicates were seeded. (L) HMDMs were primed for 4 h with 100 ng/mL LPS, followed by stimulation with 5 µM nigericin for 1 h. Cells were treated with the PP2A inhibitor okadaic acid (1 µM) 15 min before adding nigericin. Supernatants were collected and IL-1β cytokine secretion was measured using ELISA. (M) Caspase-1 was detected in HMDM cell lysates (XT) using Western blot, with one representative Western blot shown. Data in L are shown as mean + SEM from pooled results of two independent experiments with four donors, with one representative Western blot shown in M. **P < 0.01, ***P < 0.001, ****P < 0.0001.
As previously reported, LPS and nigericin activated NLRP3 and induced the classical hallmarks of inflammasome activation: secretion of IL-1β (Fig. 1B) and caspase-1 self-cleavage and secretion of the caspase 1 p20 fragment (Fig. 1H). Treatment with okadaic acid, used at 1 μM, blocked these NLRP3-dependent responses (Fig. 1 B and H). We tested three different suppliers of okadaic acid (SI Appendix, Fig. S1A) and in all three cases we measured a decrease in IL-1β secretion (SI Appendix, Fig. S1 A and B) and protection from cell death (SI Appendix, Fig. S1C). Importantly, the NLRP3-independent tumor necrosis factor (TNF) secretion was not affected (SI Appendix, Fig. S1D). Okadaic acid also blocked NLRP3 activation induced by zymosan, Pam3CSK4, and Poly(I:C) with either nigericin, MSU, or R837 stimulation (Fig. 1 C–J). Like PP2A inhibitors, PP2A deletion using siRNA reduced inflammasome-dependent IL-1β secretion (Fig. 1K) but not inflammasome-independent TNF secretion in iBMDMs (SI Appendix, Fig. S1 E and F). Thus, the PP2A-sensitive phospho-switch appears to be a broad mechanism of NLRP3 inflammasome regulation downstream of multiple TLRs in mouse BMDMs.
To investigate whether the PP2A-dependent phospho-switch is conserved in humans we induced inflammasome activation with LPS and nigericin in HMDMs as described before (21). Also in these cells we found that okadaic acid fully blocked the secretion of IL-1β (Fig. 1L) and cleavage of caspase-1 (Fig. 1M), indicating that licensing of the NLRP3 pathway by PP2A is conserved in humans.
Finally, to test whether the PP2A phospho-switch controls other inflammasomes, we analyzed AIM2 (Absent in melanoma 2) and NLRC4 (NLR family CARD domain–containing protein 4) inflammasome responses in the presence of okadaic acid. Okadaic acid was added before transfection of BMDMs with calf thymus DNA or flagellin, which activate AIM2 and NLRC4, respectively. Okadaic acid only partially reduced AIM2-mediated responses (IL-1β release and viability) (SI Appendix, Fig. S1 G and H), and it did not affect NLRC4 inflammasome response (IL-1β release and viability) (SI Appendix, Fig. S1 I and J).
PP2A ON Switch Licenses the NLRP3 Inflammasome Activity In Vivo.
To validate the above findings in vivo, we selected LB-100, a PP2A inhibitor used in clinical trials for cancer (43) and first confirmed its effects on the NLRP3 inflammasome in vitro. Similarly to okadaic acid, LB-100 inhibited all NLRP3-dependent responses (IL-1β, viability, and caspase-1 secretion) (SI Appendix, Fig. S2 A–C and E), albeit at a slower rate, as it needed a longer time (2 to 4 h) to exert effects similar to those observed with okadaic acid treatment (PP2A inhibitor 1) for 30 min. LB-100 did not affect priming of the NLRP3 inflammasome as it did not block the LPS-induced TNF secretion (SI Appendix, Fig. S2D) or the induction of pro–IL-1β expression (SI Appendix, Fig. S2E).
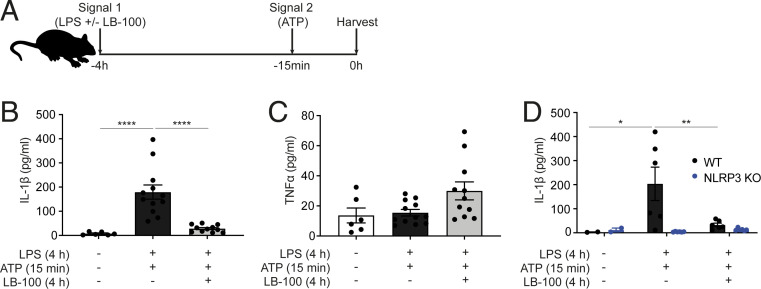
We then used the in vivo acute peritonitis model induced by LPS and adenosine 5′-triphosphate (ATP) (Fig. 2A), where LB-100 was dosed with LPS, and after 4 h, ATP was administered for 15 min. LB-100 completely blocked IL-1β secretion in the peritoneal lavage without altering TNF secretion (Fig. 2 B and C). If anything, there was a trend for increased TNF secretion, suggesting that the PP2A inhibitor LB-100 may prevent cell death (pyroptosis) when it inhibits the NLRP3 inflammasome, thereby allowing prolonged TNF release from live peritoneal cells. Importantly, we showed that IL-1β secretion in this peritonitis model is dependent on the NLRP3 inflammasome as IL-1β secretion was greatly reduced in the NLRP3 knockout (KO) mice (Fig. 2D). At this time point LB-100 did not significantly change the percentage of CD11b+ myeloid cells in the peritoneum (SI Appendix, Fig. S2 F, G, and H).
Fig. 2.
PP2A ON switch licenses the NLRP3 inflammasome activity in vivo. (A) WT or NLRP3 KO mice, 8 to 12 wk old, were injected i.p. with either PBS (control) or 1 µg LPS with or without 10 µg LB-100. After 3 h 45 min mice were injected i.p. with either PBS (control) or 200 µL of 25 mM ATP. Fifteen minutes later, mice were killed and peritoneal cavity was washed with 5 mL ice-cold PBS and 3% FBS to obtain peritoneal lavage for downstream analysis. (B) IL-1β concentration levels in the peritoneal lavage of WT mice treated with or without LB-100 was measured by ELISA. (C) TNF concentration levels in the peritoneal lavage of WT mice treated with or without LB-100 was measured by ELISA. (D) IL-1β concentration levels in the peritoneal lavage of WT and NLRP3 KO mice treated with or without LB-100 was measured by ELISA. In all panels dots represent individual mice and columns depict mean + SEM. Data shown are pooled from three independent experiments in B and C or two independent experiments in D. *P < 0.05, **P < 0.01, ****P < 0.0001.
Collectively, these data indicate that the PP2A ON switch licenses the NLRP3 inflammasome activity in vivo.
Pharmacological PP2A Activation Is Not Sufficient to Activate the NLRP3 Inflammasome Pathway.
Previous (32) and our results above suggest that to prevent unwanted activation, NLRP3 is maintained in a signaling-incompetent state through a posttranslational phospho-brake present both in mice and humans. The brake is removed by the action of the phosphatase PP2A. In order to answer the question whether PP2A activation can, on its own, activate the NLPR3 inflammasome pathway, we primed primary BMDMs with LPS (signal 1) for 4 h and stimulated them with nigericin (signal 2) for 1 h. The pharmacological PP2A activator DBK-1154 (44) was added 30 min before nigericin (SI Appendix, Fig. S3A). We found that PP2A activation did not alter IL-1β or TNF secretion (SI Appendix, Fig. S3 B and D) or cell viability (SI Appendix, Fig. S3C) in either basal or LPS + nigericin–stimulated condition. The activity of PP2A activator was confirmed using a phosphatase activity assay (SI Appendix, Fig. S3E). Together, these data suggest that PP2A acts as a licensing signal whose activity is necessary but not sufficient, on its own, to activate the NLRP3 inflammasome.
TBK1 and IKKε Kinase Act as an OFF Switch to Limit NLRP3 Inflammasome Activation.
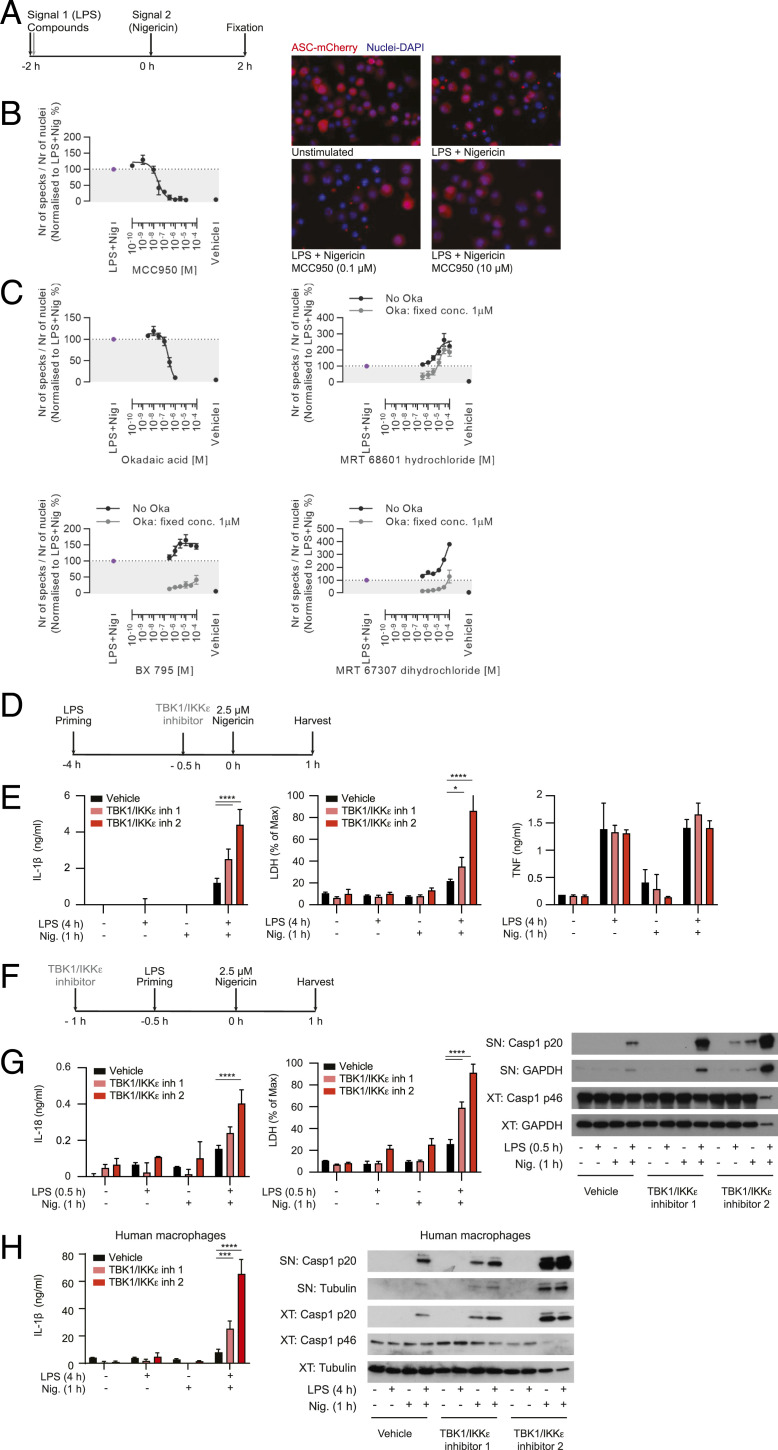
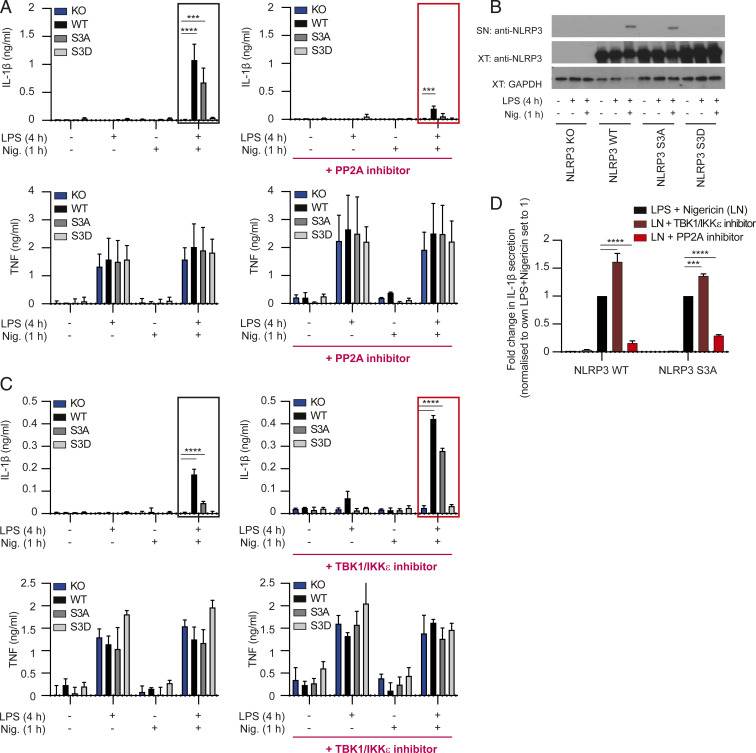
If a phosphatase controls the ON switch, a kinase must control the OFF switch. In an endeavor to identify NLRP3 pathway regulators, we unexpectedly discovered that a number of reported TBK1/IKKε inhibitors increased inflammasome activation (Fig. 3 A and C). The kinase domains of TBK1 and IKKε are closely related and most inhibitors block both of their activities. The assay that we used to carry out these studies was based on detection of fluorescent ASC-mCherry in stably transfected iBMDMs, where ASC specks are induced after cell stimulation with 1 µg/mL LPS for 2 h and 10 µM nigericin for 2 h (Fig. 3A). The image-based assay was validated using the pharmacological NLRP3 inhibitor MCC950 (also known as CRID3), which decreased ASC speck formation (Fig. 3 B, Right) in a concentration-dependent manner with an IC50 (concentration that inhibits response by 50%) of 24 nM (Fig. 3 B, Left). The PP2A inhibitor okadaic acid also inhibited NLRP3 activation by decreasing speck formation in a concentration-dependent manner with an IC50 of 0.2 µM (Fig. 3C). Surprisingly, the TBK1/IKKε inhibitors MRT 68601, BX 795, and MRT 67307 each enhanced ASC speck formation in a concentration-dependent manner (Fig. 3C). They also completely or partially reversed the okadaic acid effect on ASC speck in a concentration-dependent manner (Fig. 3C). Collectively these results suggest that the TBK1 and/or IKKε kinase play an important role in controlling the NLRP3 inflammasome OFF switch and act against the PP2A phosphatase ON switch to balance NLRP3 pathway activity.
Fig. 3.
TKB1/IKKε inhibitors increase NLRP3 inflammasome activation and reverse PP2A effects in vitro. (A) The schematic shows ASC-mCherry iBMDMs treatment with 1 µg/mL LPS in combination with compounds for 2 h and subsequently 10 µM nigericin (subsaturating dose of nigericin for iBMDMs) for 2 h, after which PFA was added. (B) ASC speck visualization and quantification after LPS and nigericin treatment in the presence of the NLRP3 inhibitor MCC950. (C) Concentration response curve of okadaic acid (Top Left) effect on ASC speck formation or concentration response curve of MRT 68601 (Top Right), BX 795 (Bottom Left), and MRT 67307 (Bottom Right) effect on ASC speck formation in the presence or absence of a fixed concentration of okadaic acid (1 µM). Data shown are mean ± SEM from pooled results of three or more (B and C) independent experiments. (D and F) Primary BMDMs were primed with 100 ng/mL LPS for 4 h (chronic priming) or 30 min (acute priming) followed by stimulation with 2.5 µM nigericin (subsaturating dose of nigericin for BMDMs) for 1 h. Cells were treated with the TBK1/IKKε inhibitor 1 (MRT 67307) or TBK inhibitor 2 (MRT 68601), each at 10 µM, for 30 min before nigericin stimulation in the chronic assay or 30 min before LPS stimulation in the acute assay. Supernatants and cell lysates were collected 1 h after nigericin addition. (E) IL-1β and TNF cytokine secretion were measured using ELISA; cell viability was measured by LDH release assay. (G) Because IL-1β is not transcriptionally induced in the acute inflammasome priming assay, IL-18 secretion was used as a readout of inflammasome stimulation. IL-18 was measured by ELISA. Caspase-1 was measured in cell supernatants and lysates using Western blot. Data are shown as mean + SD of triplicates from one experiment representative of three for E and G. (H) HMDMs were primed with 100 ng/mL LPS for 4 h followed by stimulation with 2.5 µM nigericin (subsaturating dose of nigericin for HMDMs) for 1 h. IL-1β cytokine secretion was measured using ELISA. Caspase-1 was measured in cell supernatants and lysates using Western blots. Data are shown as mean + SD of triplicates from one representative of two independent experiments with one representative Western blot. *P < 0.05, ***P < 0.001, ****P < 0.0001.
To rule out the indirect, transcription-dependent effects of prolonged TBK1/IKKε inhibition on NLRP3 inflammasome responses, we tested the compounds using the priming and activation paradigms: Compounds were either added after LPS priming or were used in an acute, transcription-independent inflammasome activation protocol. In one set of experiments, primary BMDMs were primed with LPS for 3.5 h then treated with TBK1/IKKε inhibitors (10 µM) for 30 min, before stimulation with 2.5 μM nigericin for 1 h. Nigericin was used at a subsaturating dose, to allow the detection of increased responses (Fig. 3D). In a second set of experiments, we used a well-characterized transcription-independent NLRP3 activation assay (23). We added the TBK1/IKKε inhibitors (10 µM) 30 min before LPS priming for another 30 min, followed by nigericin stimulation for 1 h (Fig. 3F). In these assays, too, TBK1/IKKε inhibitors (MRT 67307, inhibitor 1 and MRT 68601, inhibitor 2) increased the NLRP3 inflammasome responses of IL-1β, IL-18 secretion, caspase-1 cleavage, and LDH release, without affecting TNF release (Fig. 3 E and G). Results were recapitulated in primary human macrophages, HMDMs (Fig. 3H), even with signal 2 (nigericin) alone, further supporting the idea that TBK1 and/or IKKε may be a new, conserved OFF switch for the NLRP3 pathway.
We also tested whether TBK1/IKKε affected the activity of other inflammasomes such as AIM2 or NLRC4 and utilized the methodology described above, with the TBK1/IKKε inhibitor 2 added 30 min before stimulation with calf-thymus DNA or flagellin for 1 h and 4 h in iBMDMs or 4 h in BMDMs (SI Appendix, Fig. S4A). TBK/IKKε inhibitors did not affect AIM2 and NLRC4 responses (IL-1β secretion and cell death) (last two bars on the right in all panels of SI Appendix, Fig. S4 B–E). However, prolonged time required to detect AIM2 and NLRC4 inflammasomes activation in BMDMs meant prolonged exposure to TBK1/IKKε inhibitor (4 to 5 h). This led to increased basal responses to LPS alone in the presence of TBK1/IKKε inhibitor, making indirect effects of TBK1/IKKε inhibition on apoptosis, or on the autophagy pathway, a well-known regulator of the NLRP3 inflammasome, hard to control in the long-term assays.
TBK1 and IKKε Are Related Kinases That Can Compensate for Each Other’s Function in Limiting NLRP3 Activity.
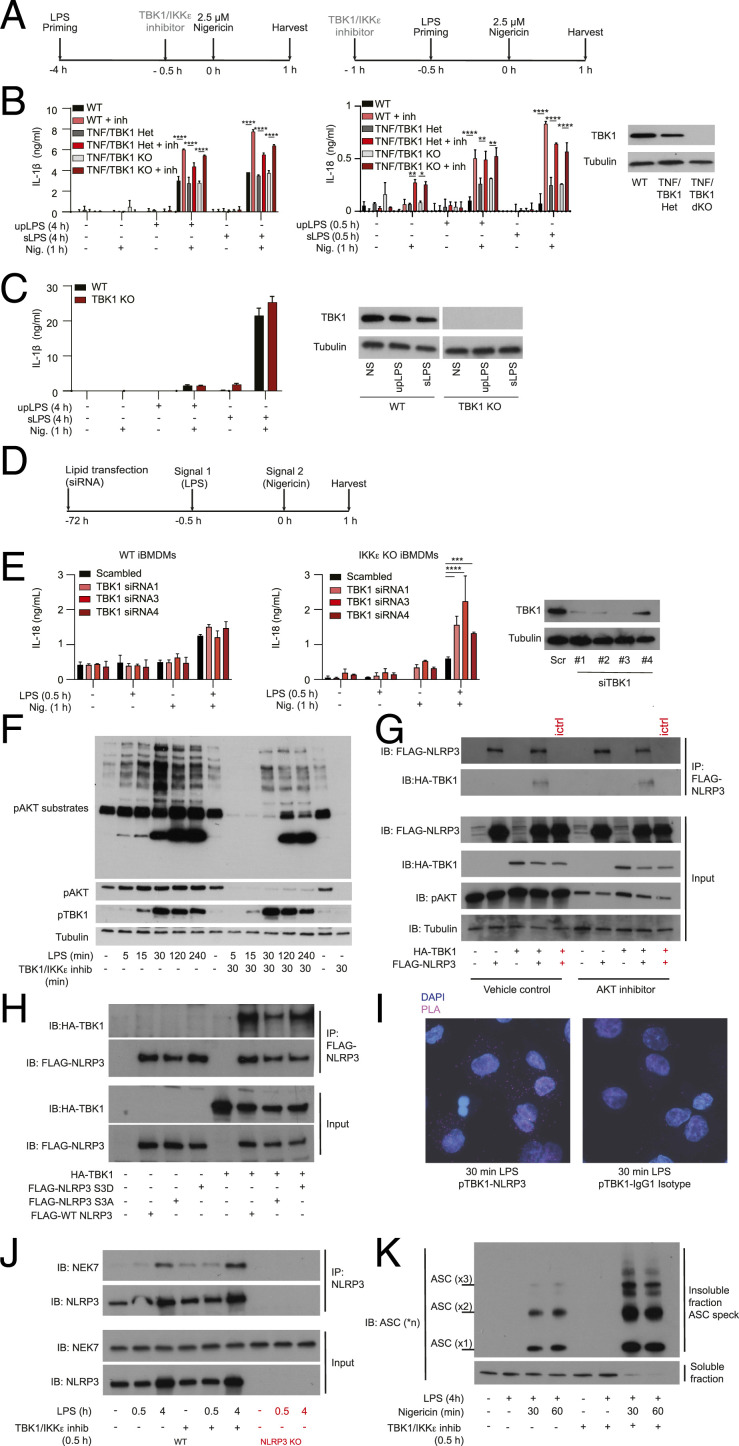
Because TBK1 kinase is closely related with the IKKε kinase, most TBK1 inhibitors also block IKKε (45, 46). Furthermore, in several signaling pathways these kinases can compensate for each other’s function (47–50). To test whether TBK1, IKKε, or both, can act as an OFF switch for the NLRP3 pathway we analyzed NLRP3 activation in macrophages lacking TBK1, IKKε, or both. TBK1 deletion in mice is lethal but can be reversed by simultaneous deletion of TNF (51, 52). So, we first analyzed NLRP3 inflammasome activation in WT, Tnf−/−;Tbk1+/− (labeled as TNF/TBK1 HET) or Tnf−/−Tbk1−/− (labeled as TNF/TBK1 KO) BMDMs. Cells were stimulated with LPS for 4 h (in chronic inflammasome priming assay) or 0.5 h (in acute inflammasome priming assay) followed by a subsaturating dose (2.5 μM) of nigericin for 1 h in the presence or absence of a TBK1/IKKε inhibitor (10 μM of MRT 68601) (Fig. 4A). We confirmed TBK1 deletion by Western blot and then measured NLRP3-dependent IL-1β and IL-18 secretion by enzyme-linked immunosorbent assay (ELISA) (Fig. 4B), NLRP3-dependent caspase-1 cleavage by Western blot (SI Appendix, Fig. S5A), and NLRP3-dependent cells death by LDH release assay (SI Appendix, Fig. S5B). We found that NLRP3 responses to either ultrapure LPS (upLPS) or less pure Sigma LPS (sLPS) and nigericin were similar between wild-type (WT) and TBK1-deficient cells (Fig. 4B, gray bars) and that NLRP3 response were similarly boosted in all cells by TBK1/IKKε inhibitors (Fig. 4B, red bars). The results were recapitulated in the undifferentiated human THP-1 monocyte line (Fig. 4C), suggesting again the conserved mechanism. Fully PMA-differentiated THP-1 macrophages were not used, because PMA differentiation was affected by TBK1 deficiency. Therefore, NLRP3 inflammasome responses are intact in cells lacking TBK1 only.
Fig. 4.
TBK1 and IKKε kinase act as an OFF switch to limit NLRP3 inflammasome activation. (A) BMDMs were primed with 100 ng/mL ultrapure LPS or crude Sigma LPS (L4391) for 4 h (chronic priming) or 30 min (acute priming) followed by stimulation with 2.5 µM nigericin (subsaturating dose for BMDMs) for 1 h. Cells were treated with the TBK1/IKKε inhibitor MRT 68601 at 10 µM for 30 min before nigericin stimulation in the chronic assay or 30 min before LPS stimulation in the acute assay. (B) IL-1β or IL-18 cytokine secretion were measured using ELISA after chronic and acute stimulation, respectively. TBK1 deletion was confirmed using Western blot. (C) Undifferentiated WT or TBK1 KO THP-1 monocytes were primed with 1 μg/mL ultrapure LPS or crude Sigma LPS (L4391) for 4 h followed by stimulation with 2.5 µM nigericin for 1 h. IL-1β secretion was measured using ELISA and TBK1 deletion was confirmed using Western blot. (D) Transfection schematic. WT or IKKε KO iBMDMs were incubated with Lipofectamine RNAiMAX transfection reagent with 10 nM TBK1 siRNA or with 10 nM scrambled siRNA for 72 h. Cells were then primed with 1 µg/mL ultrapure LPS for 30 min, followed by stimulation with 10 µM nigericin for 1 h min, after which supernatants were collected and cells lysed. (E) IL-18 secretion was measured using ELISA and TBK1 deletion was confirmed using Western blot. IL-18 data from siRNA2 were not available in all repeats so were not included in the final ELISA analysis. Data for WT and IKKε KO iBMDMs are from the same experiment, plotted in two separate panels because ELISAs was run on two separate 96-well plates due to large number of samples and conditions. Each siRNA result is thus compared to its own scrambled control. The y axes were also matched within each experiment for comparison. (F) Representative Western blot for AKT substrate, TBK1, and AKT phosphorylation after a time-course stimulation of WT BMDMs treated with 100 ng/mL LPS, in the presence or absence of 10 µM TBK1/IKKε inhibitor MRT 68601, added 30 min before LPS. (G) Coimmunoprecipitation of mouse FLAG-NLRP3 and mouse HA-TBK1 ectopically expressed in HEK293T cells using transient transfection. NLRP3 was immunoprecipitated from samples using anti-NLRP3 antibody or isotype control (ictrl). One micromolar AKT inhibitor MK2206 was added 60 min before cell lysis. (H) Coimmunoprecipitation of mouse FLAG-NLRP3 WT, S3A or S3D, and mouse HA-TBK1 ectopically expressed in HEK293T cells using transient transfection. (I) PLA analysis to detect interaction between endogenous phosho-TBK1 and NLRP3 (or isotype control) in PMA-differentiated THP1 macrophages 30 min after LPS stimulation. Representative 63× images are shown. Quantification of all cells is in SI Appendix. (J) Coimmunoprecipitation of endogenous NEK7 and NLRP3 from WT iBMDMs (or control NLRP3 KO cells) before and after LPS stimulation in the presence of a vehicle control or 10 µM TBK1/IKKε inhibitor MRT 68601 added 30 min before LPS. (K) ASC speck isolation after DSS cross-linking from iBMDMs stimulated with LPS + nigericin in the presence or absence of 10 µM TBK1/IKKε inhibitor MRT 68601, added 30 min before nigericin. In all cells 10 µM of caspase-1 inhibitor VX-765 was added 30 min before TBK1/IKKε inhibitor MRT 68601 to prevent cell death and loss of ASC specks. Data are shown as mean + SD of triplicates from one experiment representative of two for B, of three for C and E, one representative Western blot from two experiments for F, G, and J and three for H and K, and one representative PLA experiment of two for I. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To test whether in the absence of TBK1, IKKε may be compensating for TBK1 function in limiting NLRP3 pathway activity we repeated the acute inflammasome activation assay in WT and IKKε KO iBMDMs, before and after deletion of TBK1 using TBK1 siRNA. Cells were primed with LPS for 0.5 h and then stimulated with nigericin for 1 h. Inflammasome response was measured by IL-18 secretion. When compared to WT cells, Ikkε−/− (IKKε KO) iBMDMs always had reduced IL-18 secretion in response to LPS + nigericin, suggesting a requirement for IKKε in the priming of NLRP3 pathway. However, TBK1 deletion by siRNA (Fig. 4 D and E) in IKKε KO iBMDMs, but not in WT cells, boosted the IL-18 secretion and NLRP3-dependent caspase-1 cleavage (SI Appendix, Fig. S5C). Thus, we conclude that IKKε likely plays two roles in NLRP3 pathway, one in priming of NLRP3 activation and the other in switching off the excessive NLRP3 activity. The latter role IKKε shares with TBK1.
AKT was recently described as an OFF switch kinase for NLRP3-serine 5 (serine 3 in mice) (38). Because AKT is the best-known downstream target of TBK1, we tested whether TBK1 inhibition affects AKT activity in BMDMs. Data revealed that in BMDMs stimulated with LPS the TBK1/IKKε inhibitor did not prevent TBK1 activation at position serine172, as expected (45), but it almost completely abrogated phosphorylation of TBK1 target, AKT, and many of the downstream AKT phospho-substrates (Fig. 4F). Thus, TBK1 may limit NLRP3 activity, at least in part, by controlling the activity of AKT.
To better understand the mechanism of NLRP3 regulation by TBK1, we tested whether TBK1 can interact with NLRP3 in cells and, if so, whether the interaction is dependent on AKT activity and the AKT-substrate site, NLRP3-S3. We first ectopically expressed hemagglutinin (HA)–tagged mouse TBK1 with FLAG-tagged mouse NLRP3 in HEK293T cells and analyzed TBK1-NLRP3 interaction using coimmunoprecipitation experiments in the presence or absence of AKT inhibitor (Fig. 4G). Data revealed that, when overexpressed, TBK1 binds to NLRP3 and that binding is independent of AKT activity. AKT inhibitor reduced AKT phosphorylation in HEK293T cells, as expected, but it did not affect TBK1–NLRP3 binding (Fig. 4G).
To test whether TBK1–NLRP3 interaction is dependent on the AKT-substrate site NLRP3-serine 3, we then expressed TBK1 in HEK293T cells with either WT NLRP3 or NLRP3-serine 3 mutants, S3A and S3D. We predicted that in these constructs S3A would mimic dephosphorylated and active NLRP3, while S3D would mimic constitutively phosphorylated and inactive NLRP3. Data revealed that, when overexpressed, TBK1 binds to NLRP3, either WT or serine 3 mutants (Fig. 4H), indicating that the TBK1–NLRP3 interaction likely occurs at a site distinct from serine 3. Thus, TBK1 binding to NLRP3 is independent of AKT and the AKT-target site, NLRP3-serine 3.
We were unable to coimmunoprecipitate the endogenous TBK1 and NLRP3 in macrophages, suggesting that interaction is transient, or indirect. However, we were able to use proximity ligation assay (PLA) and microscopy to capture the interaction between active, endogenous phospho-TBK1 and endogenous NLRP3 in LPS-primed THP1 macrophages, as early as 30 min upon LPS stimulation (Fig. 4I and SI Appendix, Fig. S6A). The interaction was not detected in unstimulated cells or in cells stained with isotype control antibodies. Furthermore, we detected the strongest PLA colocalization signal 30 to 60 min upon LPS stimulation (SI Appendix, Fig. S6 A and B), corresponding well to the kinetic of TBK1 activation in LPS-treated macrophages (Fig. 4F). These results are consistent with a recent study that reported the presence of phosho-TBK1 in the NLRP3 signaling complex using proteomics and microscopy (53). Thus, TBK1 interacts with the NLRP3 complex in macrophages and limits its activity. As TBK1 activation is induced downstream of TLRs that prime NLRP3 expression, a model emerges that the same signals that prime NLRP3 expression also introduce brakes that prevent its premature activation without a bona fide signal 2.
To further define how TBK1 and IKKε control NLRP3 activity, we tested whether they regulate the interaction of NLRP3 with the upstream activating kinase NEK7 after priming signal 1, or downstream formation of ASC specks upon detection of activating signal 2. We found that NLRP3 interaction with NEK7 was similar in control iBMDMs and iBMDMs treated with TBK1/IKKε inhibitor (Fig. 4J). However, the formation of ASC specks was markedly increased in response to LPS + nigericin as early as 30 min postactivation in the presence of TBK1/IKKε inhibitor (Fig. 4K), further supporting the increased caspase-1 activation, cell death, and cytokine secretion response in the absence of TBK1/IKKε brake. Because the activity of AIM2 and NLRC4 inflammasomes, which share the downstream effectors with NLRP3, was not equally boosted by TBK1/IKKε inhibition (SI Appendix, Fig. S4), data suggest that TBK1/IKKε brake controls either the NLRP3 itself or its direct interacting partner.
Serine 3 (Serine 5 in Humans) Is an Important Phospho-Switch for NLRP3 but Is Not the Sole Site Controlled by Either PP2A or TBK1/IKKε.
TBK1 activity in the NLRP3 pathway can be in part explained by its control over AKT. AKT inhibits NLRP3 through phosphorylation on NLRP3-serine 5 in humans (serine 3 in mice). Mutagenesis of NLRP3-serine 3 makes the pathway insensitive to further AKT control, suggesting that serine 3 is indeed the main site of AKT action (38). To genetically confirm that serine 3 is the phospho-switch site in NLRP3 pathway and to test whether this is also the sole site under TBK1/IKKε control, we subcloned mouse WT NLRP3, nonphosphorylable S3A or phospho-mimetic S3D mutants into retroviral vectors. We expressed these constructs in Nlrp3-deficient (NLRP3 KO) mouse bone marrow progenitors prior to differentiating cells into mature BMDMs. All constructs expressed equally well in NLRP3 KO cells, as measured by the expression of green fluorescent protein (GFP) encoded by the same plasmid carrying the NLRP3 mutants (SI Appendix, Fig. S7). In agreement with our hypothesis and with the findings from immortalized lines (32), WT and nonphosphorylable/active S3A mutants transduced in NLRP3 KO macrophages enabled LPS/nigericin–induced inflammasome-dependent IL-1β secretion and NLRP3 release from cells with active inflammasome, while the phospho-mimetic/inactive S3D mutant was completely unresponsive to inflammasome stimulation (Fig. 5 A, Top Left, and B), despite similar inflammasome-independent TNF secretion in BMDMs expressing each of the three constructs (Fig. 5 A, Bottom Left). These data confirm serine 3 as an important phospho-switch site for the NLRP3 pathway in primary BMDMs.
Fig. 5.
NLRP3 serine 3 is not the sole site licensed for activation by either PP2A or TBK1/IKKε. (A) NLPR3 KO bone marrow was retrovirally transduced to express WT or serine 3 NLRP3 mutants. Resulting BMDMs were primed for 4 h with 100 ng/mL LPS, followed by stimulation with 5 µM nigericin for 1 h. Cells were treated with vehicle control (Left) or okadaic acid used at 1 μM (Right) for 30 min before nigericin stimulation. Supernatants were collected and IL-1β and TNF cytokine secretion were analyzed by ELISA. Data for vehicle and inhibitor treatments were from the same experiment, plotted in two separate panels because ELISA was run on two 96-well plates due to the large number of samples and conditions. Responses of NLRP3 mutants (in the absence or presence of inhibitors) were analyzed relative to their internal NLRP3 WT control. The y axes were also matched within each experiment for comparison. Data are shown as mean + SE of mean of pooled results from three independent experiments for vehicle control and two experiments for okadaic acid. (B) NLRP3 protein was detected in supernatants (SN) and cell lysates (XT) from transduced NLRP3 KO BMDMs stimulated as in A. One representative blot is shown. (C) NLRP3 KO BMDMs expressing WT or serine 3 NLRP3 mutants were prepared as in A. Cells were then primed for 4 h with 100 ng/mL LPS, followed by stimulation with 2.5 µM nigericin for 1 h. A subsaturating concentration of nigericin was used here to allow detection of increased inflammasome responses in TBK1/IKKε inhibitor-treated cells. Cells were treated with vehicle control (Left) or 10 µM TBK1/IKKε inhibitor MRT 68601 (Right) for 30 min before nigericin stimulation. Supernatants were collected and IL-1β and TNF cytokine secretion were determined using ELISA as in A. Data are shown as mean + SD or triplicates from one representative of two independent experiments. (D) Key LPS + nigericin–treated samples from A and C were reanalyzed on the same plate to allow statistical comparison of cells treated in the absence or presence of TBK1/IKKε or PP2A inhibitors. Data are shown as normalized mean + SD of triplicates from one representative of two independent experiments. ***P < 0.001, ****P < 0.0001.
If serine 3 is the sole site in the NLRP3 pathway controlled by PP2A, or TBK1/IKKε, then the nonphosphorylable/active S3A mutant should make the NLRP3 pathway insensitive to PP2A or TBK1/IKKε inhibitors. Surprisingly, we found that, unlike for AKT (38), this was not the case for either TBK1/IKKε or PP2A, as the BMDMs expressing S3A were as sensitive to PP2A (okadaic acid at 1 μM) and TBK1/IKKε inhibitor (MRT 68601 at 10 μM) as WT cells (Fig. 5 A, Top, Fig. 5 C, Top, and Fig. 5D). Both inhibitors affected inflammasome-dependent IL-1β, but not inflammasome-independent TNF secretion (Fig. 5 A, Bottom, and Fig. 5 C, Bottom). Together, the data suggest that serine 3 is actually not the sole site in the pathway regulated by either the TBK1/IKKε or PP2A, as both the kinases and the phosphatase retained their control over NLRP3 activity even when serine 3 is mutated.
Discussion
Activation of the NLRP3 inflammasome can be beneficial during infection and vaccination. Nonetheless, when NLRP3 activity is uncontrolled and chronic, it becomes detrimental and contributes to inflammation-driven pathology in several diseases, such as Alzheimer’s disease and atherosclerosis. In healthy individuals, therefore, there must exist a licensing mechanism that prevents unwanted NLRP3 inflammasome responses. Here, we characterized one such mechanism. A recent study (32) identified PP2A as the ON switch for NLRP3 in iBMDMs. The report by Stutz et al. (32) raised several important questions: How conserved is the ON switch, does it work in vivo, and which kinase controls the OFF switch? In addressing these questions, we first discovered that the PP2A ON switch is conserved in mouse and human macrophages, downstream of most known NLRP3 priming and activating signals, and that it works in vivo to control NLRP3-driven inflammation. We then identified that TBK1 and IKKε act as a novel OFF switch for NLRP3 pathway. Using pharmacological and genetic approaches we showed that blockade or deletion of both TBK1 and IKKε, but neither kinase alone, enhance the responses downstream of the NLRP3 inflammasome activation. In addition, TBK1/IKKε inhibitors counteract the effects of PP2A inhibition on inflammasome activity, suggesting that TBK1 and IKKε work to oppose the action of the PP2A ON switch. Mechanistically, we find that TBK1/IKKε may be limiting NLRP3 activity, at least in part, by controlling the activity of another known NLRP3 OFF switch kinase, AKT. However, TBK1/IKKε control also goes beyond AKT, as TBK1 interacts with NLRP3 and acts at a site distinct from NLRP3-serine 3, previously reported to be under the AKT control. Mutagenesis of NLRP3 confirms serine 3 as an important phospho-switch site but surprisingly reveals that, unlike for AKT, this is not the sole site regulated by either TBK1/IKKε or PP2A, as all retain the control over the NLRP3 pathway even when serine 3 is mutated. Together, we propose that TBK1 and IKKε act as an OFF switch for the NLRP3 pathway, that they can compensate for each others function in this process, and that they can counteract the PP2A ON switch phosphatase to balance NLRP3 activity.
TBK1 and IKKε are serine/threonine kinases activated downstream of many TLRs, which also prime the NLRP3 pathway. Thus, the OFF brake is likely introduced by TLRs through TBK1/IKKε at the time of NLRP3 expression and priming to prevent spontaneous NLRP3 activation in the absence of signal 2. The OFF brake is then only removed by the action of the PP2A phosphatase upon the detection of the inflammasome-activating signal 2, such as the bacterial pore-forming toxin nigericin or the endogenous danger signal, ATP. Interestingly, TBK1 and IKKε introduce a similar posttranslational phospho-brake on RIPK1, to block other death pathways, apoptosis (54), and necroptosis (47). Together these findings point to a conserved mechanism of cell survival, induced downstream of various TLRs, through TBK1 and IKKε, that allows the initiation of inflammatory responses, by putting the brakes on cell death pathways. These brakes are only removed upon the loss of cellular homeostasis, licensing the death of a compromised cell. Interestingly, another broader inhibitor of PP1 and PP2A phosphatases, calyculin A, was reported to block AIM2 and NLRC4 responses (55), suggesting that phospho-switches exist in other inflammasomes as well but are likely regulated by distinct phosphatases, such as PP1.
The exact targets in the NLRP3 pathway phosphorylated by TBK1 or IKKε will need future evaluation. Proteomics analyses of iBMDMs proposed serine 3 of NLRP3 (serine 5 in humans) as the PP2A-sensitive site. Using an NLRP3-S3D mutant to mimic phosphorylated and inactive NLRP3, we genetically confirmed that serine 3 is one phospho-switch for the NLRP3 pathway in mice. A recent study, published during the course of this work, identified AKT as the direct kinase for serine 5 (serine 3 in mice) (38). AKT is the best-known target of TBK1, and we found that in BMDMs TBK1 inhibition blocked AKT activation and the phosphorylation of downstream AKT substrates, including, presumably, NLRP3-serine 3. Thus, we propose that, at the time of priming, TLRs activate TBK1 and its main downstream substrate, AKT, to put a brake on NLRP3 activation. The phospho-brake is then removed by the action of PP2A only upon detection of signal 2 and the loss of cellular homeostasis.
If the serine 3 residue was the sole site controlled by PP2A, or TBK1/IKKε, then our next hypothesis was that the nonphosphorylable/active S3A mutant should make the NLRP3 pathway insensitive to PP2A or TBK1/IKKε inhibitors. However, this was not the case as both WT and S3A mutant NLRP3 were still sensitive to both PP2A and TBK1 inhibition. Thus, while serine 3 is an important phospho-switch this is not the sole site regulated by either TBK1/IKKε and PP2A in NLRP3 pathway. In support of this conclusion, we found that TBK1 interacts with NLRP3 at a site distinct from serine 3 and maintains this interaction even when AKT activity is inhibited. Future proteomic analyses of NLRP3 complexes in the presence or absence of PP2A or TBK1/IKKε inhibitors should identify the TBK1 substrate in the NLRP3 complex. It was suggested that cellular trafficking of ASC depends on PP2A inhibition of ASC-associated kinase IKKα, and this may be an additional PP2A-sensitive step in the NLRP3 pathway (56).
In most assays we used TBK1/IKKε inhibitors for 30 min only, to focus the study on direct, transcription-independent control of NLRP3 activity. This also allowed us to separate the role of TBK1/IKKε in TLR signaling, i.e., NLRP3 pathway priming, from their role in also limiting the premature NLRP3 activity. However, it is important to note that in the long term, TBK1 is also a major inducer of type I interferons and autophagy (57, 58), both of which are well-known negative regulators of NLRP3 inflammasome protein expression and availability. Thus, in a larger view, the direct posttranslational phospho-switches, like the one described here, likely work in combination with other, transcription-dependent mechanisms to limit NLRP3 availability and activity.
Finally, in the absence of NLRP3 activating signal 1 or 2, the nonphosphorylable/active NLRP3-S3A mutant did not show spontaneous pathway activation. These data support results from other mutagenesis studies (30, 32, 38) and a general model that multiple posttranslational sites control NLRP3 activation. Interestingly, when mutated they all appear nonredundant in the control of pathway activity. Hence, we hypothesize that NLRP3 PTMs are interdependent, like they are in the nuclear factor κB pathway, and likely work in a linear manner to control different steps of NLRP3 activation such as NLRP3 protein stabilization, trafficking to the activation site, activation-induced conformational change, ATP binding, and oligomerization.
Multiple targeting strategies to control NLRP3 activity at different points in the pathway will need to be developed, as acquired resistance to a single therapeutic target is a common issue. Mice expressing dominant active, patient-associated NLRP3 mutations taught us a lot about what happens when the NLRP3 pathway is constitutively turned on (59). Here we used an NLRP3-dependent peritonitis model, induced by LPS and ATP, as proof-of-principle evidence that the PP2A-sensitive phospho-switch works in vivo and can control NLRP3-driven inflammation. Drugs that modify PP2A activity are currently in trials for cancer (43, 60). If such drugs turn out to inhibit or activate the NLRP3 inflammasome pathway in a disease setting, this will be an important mechanism of action that we need to understand. It is critical to characterize the endogenous mechanisms (61), like the one we described here, by which unwanted NLRP3 inflammasome activation in healthy people is prevented, and to use this knowledge to turn the pathway ON or OFF, as needed, when designing knowledge-based therapies or vaccines.
Materials and Methods
Mice and Cells.
C57BL/6 mice were purchased from Charles River. NLRP3 KO (B6.Nlrp3tm1Vmd) mice were obtained from Kevin Maloy, University of Glasgow, Glasgow, Scotland, and were originally from Vishva Dixit, Genentech, San Francisco, CA. In-house-bred C57BL/6 mice were used as a source of bone marrow cells for generation of bone marrow–derived macrophages in vitro. All mice were housed and bred under specific pathogen-free conditions, and all studies were performed in accordance with the ethical standards approved by the Home Office and the University of Oxford. WT and IKKε KO iBMDMs were generated by Jonathan Kagan, Harvard University, Cambridge, MA. WT, Tnf−/−;Tbk1+/− (TNF/TBK1 HET) or Tnf−/−Tbk1−/− (TNF/TBK1 KO) bone marrow was from Cevayir Coban and Michelle Sue Jann Lee, University of Tokyo, Tokyo, Japan.
In Vivo Peritonitis Model.
Either male or female WT or NLRP3 KO mice between 8 and 12 wk old were injected intraperitoneally (i.p.) with either phosphate-buffered saline (PBS) (Fisher, 10010023) or 1 µg LPS (Sigma, L4391), with or without 10 µg LB-100 (Cambridge Bioscience, L0400). Three hours and 45 min later mice were injected i.p. with either PBS (control) or 200 µL of 25 mM ATP (Sigma, A2383). Four hours after LPS injection (and 15 min after ATP injection), mice were killed by CO2 asphyxiation and their peritoneal cavities were washed with 5 mL ice-cold PBS containing 3% fetal bovine serum (FBS) (Fisher, 16000044). The washing liquid was spun down at 600 × g for 5 min to remove the cells and the remaining peritoneal lavage was collected and stored at −20 °C for subsequent cytokine measurement by ELISA.
Flow Cytometry.
For staining with extracellular markers, cells were first treated with Fc block (Biolegend, 101320) in PBS (Fisher, 10010023) with 3% FBS for 30 min. Then, the following antibodies were added: CD3-APC Cy7 (Biolegend, 100221), CD19-APC Cy7 (Biolegend, 115530), CD11c-APC (Biolegend, 117309), CD11b-BV785 (Biolegend, 101243), and live-dead dye (Fisher, L10119), and cells were further incubated on ice in the dark for another 30 min. Cells were then washed with PBS and fixed with 4% paraformaldehyde (PFA) (Fisher, 28908) for 15 min at room temperature. Samples were acquired on a LSR II cytometer (BD) and data were analyzed using FlowJo software.
Generation of Primary Mouse BMDMs.
BMDMs were generated by differentiating them from total mouse bone marrow for 7 d with recombinant macrophage colony-stimulating factor (M-CSF) (50 ng/mL, Immunotools, 11343118). Cells were cultured in complete macrophage medium consisting of RPMI (Fisher, 21875091) with 10% FBS (Gibco, certified low endotoxin, 1600044), 1× Pen/Strep/Glutamine (Fisher, 10378016), and 10 to 25 mM Hepes (Fisher, 15630056) at 37 °C with 5% CO2 and supplemented with fresh media containing 50 ng/mL M-CSF on day 5. After day 7 of differentiation, cells were replated and on day 8 cells were stimulated for inflammasome experiments.
Generation of HMDMs.
Peripheral blood mononuclear cells were isolated using Ficoll gradient from healthy donors from NHS Oxford blood bank (REC 11/H0711/7). CD14+ magnetic beads (Biolegend, 8802-6834-74) were used to positively select monocytes. CD14+ cells were differentiated into macrophages by culturing them for 7 d with M-CSF (100 ng/mL, Immunotools, 11343118). Cells were cultured in IMDM (Fisher, 21980032) with 10% FBS (Gibco, certified low endotoxin, 1600044) and 1× Pen/Strep/Glutamine (Fisher, 10378016) at 37 °C with 5% CO2 and supplemented with fresh media containing 100 ng/mL M-CSF on day 5. After day 7 of differentiation, cells were replated and on day 8 cells were stimulated for inflammasome experiments.
Culturing of the ASC-mCherry iBMDM Line.
ASC-mCherry reporter stably transfected mouse iBMDM cells were a kind gift from David Brough, University of Manchester, Manchester, United Kingdom, and described previously (62). Cells were grown in culture media made of Dulbecco’s modified Eagle’s medium (Thermo Fisher, 31966021), 10% FBS (Fisher, 31966021), and 1% penicillin/streptomycin at 37 °C/5% CO2. They were trypsinized with TrypLE (ThermoFisher 12605010) for 5 min and centrifuged at 1,000 × g. Cell count and viability were measured using a cell counter and cells seeded for the experiments as described in the relevant sections.
Retroviral Transduction of Primary BMDMs.
Retroviral particles for transduction were produced by transfecting the Platinum-E (PlatE) retroviral packaging cell line (eBioscience, RV-101) with a pMIGRMCS retroviral packaging plasmid expressing GFP, as well as FLAG-tag or a FLAG-tagged WT NLRP3, S3A NLRP3, or S3D NLRP3 constructs. Transfection media was replaced after 24 h with RPMI (Fisher, 21875091) containing 10% FBS (Gibco, certified low endotoxin, 1600044) and 1× Pen/Strep/Glutamine (Fisher, 10378016) and PlatE cells were moved to 32 °C. Supernatants containing retroviral particles were collected after 24 h, filtered through 0.45-µm low-protein-binding filters (Merck Millipore, SLHVR33S), and mixed with 250 µL transfection mix (final: 20 mM Hepes, 6 µg/mL polybrene, and 50 ng/mL M-CSF). Bone marrow cells from NLRP3 KO mice were plated 1 d earlier with 50 ng/mL M-CSF to initiate proliferation of myeloid progenitors. On the day of transduction, bone marrow cells were collected and counted and 10 × 106 cells per construct were resuspended in the virus-containing supernatant mix. The cells containing each construct were seeded in four wells of non–tissue-culture-treated six-well plates at 2.5 × 106 cells per well with 2.5 mL viral supernatant. Cells were spinfected at 1,000 × g at 32 °C for 2 h. Media was topped up and supplemented with another 2 mL of RPMI (Fisher, 21875091) with 10% FBS (Gibco, certified low endotoxin, 1600044), 1× Pen/Strep/Glutamine (Fisher, 10378016), and 50 ng/mL M-CSF (50 ng/mL, Immunotools, 11343118) and incubated overnight at 37 °C, 5% CO2. The same spinfection procedure was repeated on day 2 of bone marrow differentiation. The medium was replaced on day 4 with fresh medium containing 50 ng/mL M-CSF. Transduced bone marrow cells were collected on day 6 of differentiation and aliquots were analyzed by flow cytometry (BD Canto, BD Biosciences) for transduction efficiency by GFP expression. The remaining transduced BMDMs were seeded in tissue-culture-treated 96-well plates (Corning, 3595) at a density of 1 × 106 cells/mL and incubated for adherence overnight. On day 7, the inflammasome activations assays were conducted as described below.
Additional methods are provided as SI Appendix.
Supplementary Material
Acknowledgments
We thank Professor Kevin Maloy for the NLRP3 KO mice and Professor Sebastian Joyce for helpful discussion during the project design and for editing the manuscript. We thank Professor David Brough from the University of Manchester for the iBMDM-ASC cell line. We thank Dr. Val Millar and Dr. Daniel Ebner from the Target Discovery Institute at University of Oxford for providing access to high-content imaging instrumentation. J.S.B. is supported by a Kennedy Trust KTRR start-up fellowship (KENN 15 16 06) and a Medical Research Council New Investigator Grant (MR/S000623/1). F.A.F. and S.D. are supported by Kennedy Trust KTPS Studentships. The work carried out in the Oxford Drug Discovery Institute was supported by Alzheimer’s Research UK Grant ARUK-2015DDI-OX.
Footnotes
Competing interest statement: M.O. and Mount Sinai School of Medicine have filed a patent on the PP2A activating compound used in SI Appendix, Fig. S3.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009309118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Lamkanfi M., Dixit V. M., Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Evavold C. L., Kagan J. C., How inflammasomes inform adaptive immunity. J. Mol. Biol. 430, 217–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seydoux E., et al., Effective combination adjuvants engage both TLR and inflammasome pathways to promote potent adaptive immune responses. J. Immunol. 201, 98–112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marty-Roix R., et al., Identification of QS-21 as an inflammasome-activating molecular component of saponin adjuvants. J. Biol. Chem. 291, 1123–1136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann S., et al., Activation of the NLRP3 inflammasome is not a feature of all particulate vaccine adjuvants. Immunol. Cell Biol. 92, 535–542 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Detienne S., et al., Central role of CD169+ lymph node resident macrophages in the adjuvanticity of the QS-21 component of AS01. Sci. Rep. 6, 39475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leroux-Roels G., et al., Vaccine adjuvant systems containing monophosphoryl lipid A and QS-21 induce strong humoral and cellular immune responses against hepatitis B surface antigen which persist for at least 4 years after vaccination. Vaccine 33, 1084–1091 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Swanson K. V., Deng M., Ting J. P., The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broz P., Dixit V. M., Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Youm Y. H., et al., Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 18, 519–532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heneka M. T., et al., NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature 493, 674–678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon R., et al., Inflammasome inhibition prevents α-synuclein pathology and dopaminergic neurodegeneration in mice. Sci. Transl. Med. 10, eaah4066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duewell P., et al., NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booshehri L. M., Hoffman H. M., CAPS and NLRP3. J. Clin. Immunol. 39, 277–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinarello C. A., Simon A., van der Meer J. W., Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat. Rev. Drug Discov. 11, 633–652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauernfeind F. G., et al., Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183, 787–791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franchi L., Eigenbrod T., Núñez G., Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 183, 792–796 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haneklaus M., O’Neil J. D., Clark A. R., Masters S. L., O’Neill L. A. J., The RNA-binding protein tristetraprolin (TTP) is a critical negative regulator of the NLRP3 inflammasome. J. Biol. Chem. 292, 6869–6881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer F. A., Chen K. W., Bezbradica J. S., Posttranslational and therapeutic control of gasdermin-mediated pyroptosis and inflammation. Front. Immunol. 12, 661162 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauernfeind F., Niepmann S., Knolle P. A., Hornung V., Aging-associated TNF production primes inflammasome activation and NLRP3-related metabolic disturbances. J. Immunol. 197, 2900–2908 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Bezbradica J. S., Coll R. C., Schroder K., Sterile signals generate weaker and delayed macrophage NLRP3 inflammasome responses relative to microbial signals. Cell. Mol. Immunol. 14, 118–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGeough M. D., et al., TNF regulates transcription of NLRP3 inflammasome components and inflammatory molecules in cryopyrinopathies. J. Clin. Invest. 127, 4488–4497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juliana C., et al., Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J. Biol. Chem. 287, 36617–36622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Py B. F., Kim M. S., Vakifahmetoglu-Norberg H., Yuan J., Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell 49, 331–338 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Kawashima A., et al., ARIH2 ubiquitinates NLRP3 and negatively regulates NLRP3 inflammasome activation in macrophages. J. Immunol. 199, 3614–3622 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Song H., et al., The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat. Commun. 7, 13727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Y., et al., Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 160, 62–73 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Palazón-Riquelme P., et al., USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO Rep. 19, e44766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren G., et al., ABRO1 promotes NLRP3 inflammasome activation through regulation of NLRP3 deubiquitination. EMBO J. 38, e100376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song N., et al., NLRP3 phosphorylation is an essential priming event for inflammasome activation. Mol. Cell 68, 185–197.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Spalinger M. R., et al., NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J. Clin. Invest. 126, 4388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stutz A., et al., NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J. Exp. Med. 214, 1725–1736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo C., et al., Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity 45, 802–816 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z., et al., Protein kinase D at the Golgi controls NLRP3 inflammasome activation. J. Exp. Med. 214, 2671–2693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mortimer L., Moreau F., MacDonald J. A., Chadee K., NLRP3 inflammasome inhibition is disrupted in a group of auto-inflammatory disease CAPS mutations. Nat. Immunol. 17, 1176–1186 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Barry R., et al., SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat. Commun. 9, 3001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He M., et al., An acetylation switch of the NLRP3 inflammasome regulates aging-associated chronic inflammation and insulin resistance. Cell Metab. 31, 580–591.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao W., et al., AKT regulates NLRP3 inflammasome activation by phosphorylating NLRP3 serine 5. J Immunol. 205, 2255–2264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muñoz-Planillo R., et al., K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J., Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Groß C. J., et al., K+ efflux-independent NLRP3 inflammasome activation by small molecules targeting mitochondria. Immunity 45, 761–773 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Swingle M., Ni L., Honkanen R. E., Small-molecule inhibitors of ser/thr protein phosphatases: Specificity, use and common forms of abuse. Methods Mol. Biol. 365, 23–38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung V., et al., Safety, tolerability, and preliminary activity of LB-100, an inhibitor of protein phosphatase 2A, in patients with relapsed solid tumors: An open-label, dose escalation, first-in-human, phase I trial. Clin. Cancer Res. 23, 3277–3284 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Sangodkar J., et al., Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. J. Clin. Invest. 127, 2081–2090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark K., Plater L., Peggie M., Cohen P., Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: A distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 284, 14136–14146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark K., et al., Novel cross-talk within the IKK family controls innate immunity. Biochem. J. 434, 93–104 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Lafont E., et al., TBK1 and IKKε prevent TNF-induced cell death by RIPK1 phosphorylation. Nat. Cell Biol. 20, 1389–1399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan Y., Kagan J. C., Innate immune signaling organelles display natural and programmable signaling flexibility. Cell 177, 384–398.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hemmi H., et al., The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199, 1641–1650 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balka K. R., et al., TBK1 and IKKε act redundantly to mediate STING-induced NF-κB responses in myeloid cells. Cell Rep. 31, 107492 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Bonnard M., et al., Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 19, 4976–4985 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishii K. J., et al., TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451, 725–729 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Vayttaden S. J., et al., IRAK1-mediated coincidence detection of microbial signals licenses inflammasome activation. bioRxiv [Preprint] (2019). https://www.biorxiv.org/content/10.1101/2019.12.26.888776v1 (Accessed 25 August 2021).
- 54.Xu D., et al., TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell 174, 1477–1491.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luheshi N. M., Giles J. A., Lopez-Castejon G., Brough D., Sphingosine regulates the NLRP3-inflammasome and IL-1β release from macrophages. Eur. J. Immunol. 42, 716–725 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin B. N., et al., IKKα negatively regulates ASC-dependent inflammasome activation. Nat. Commun. 5, 4977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herhaus L., TBK1 (TANK-binding kinase 1)-mediated regulation of autophagy in health and disease. Matrix Biol. 100–101, 84–98 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Shi J. H., Xie X., Sun S. C., TBK1 as a regulator of autoimmunity and antitumor immunity. Cell. Mol. Immunol. 15, 743–745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brydges S. D., et al., Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity 30, 875–887 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clark A. R., Ohlmeyer M., Protein phosphatase 2A as a therapeutic target in inflammation and neurodegeneration. Pharmacol. Ther. 201, 181–201 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang Z., Damianou A., Di Daniel E., Kessler B. M., Inflammasome activation controlled by the interplay between post-translational modifications: emerging drug target opportunities. Cell Commun. Signal. 19, 23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daniels M. J., et al., Fenamate NSAIDs inhibit the NLRP3 inflammasome and protect against Alzheimer’s disease in rodent models. Nat. Commun. 7, 12504 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.