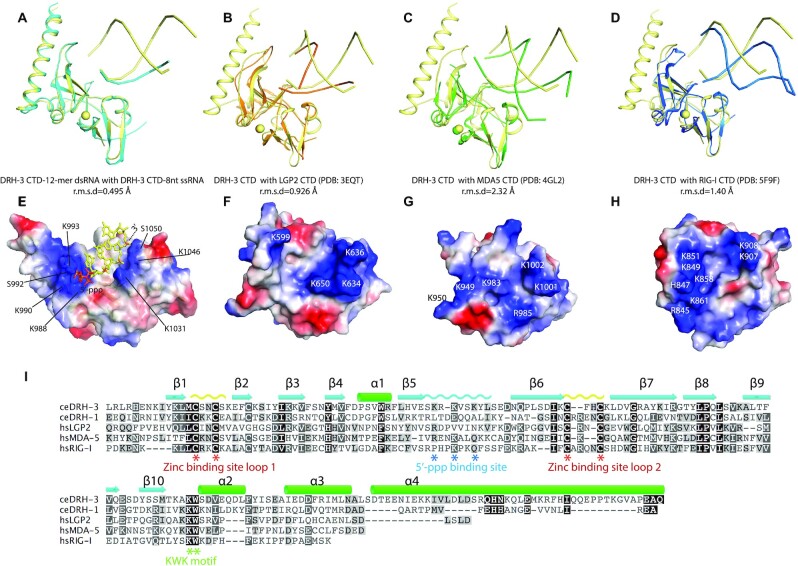
Figure 3.
Comparison of C-terminal domain of DRH3 and RLRs. (A) Superposition of DRH-3 CTD structures bound to the 5′-ppp 12-mer dsRNA and the 5′-ppp 8-mer ssRNA. (B) Superposition of the RNA-bound DRH-3 CTD (yellow) and LGP2 CTD (orange, PDB code: 3EQT). (C) Superposition of the RNA-bound DRH-3 CTD (yellow) and MDA5 CTD (green, PDB code: 4GL2). (D) Superposition of the RNA-bound DRH-3 CTD (yellow) and RIG-I CTD (orange, PDB code: 3EQT). Polypeptide chains are shown as cartoons. Structural superimposition was done by pairwise alignment and represented by Pymol (98). (E) Electrostatic charge at solvent-accessible surfaces of dsRNA-bound DRH-3 CTD. Positively charged surfaces are colored blue and negatively charged surfaces are red. Key residues involved in the RNA binding were labeled. (F–H) Electrostatic charge at solvent-accessible surfaces of LGP2 CTD, MDA5 CTD and RIG-I CTD. Each CTD is shown in the same orientation as in Figure 3A–C. Positively charged surfaces are colored in blue and negatively charged surfaces are colored in red. The key charged residues are indicated. (I) Alignment of amino acid sequences of DRH and RLR CTDs. Sequences were aligned using the program ClustalW and visualized using Geneious software. The conservation of the amino acids is indicated by colors.