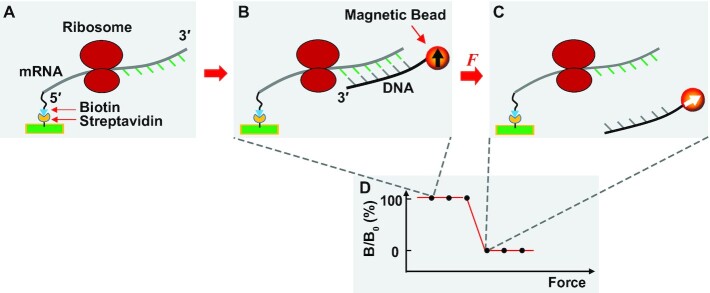
Figure 1.
A general scheme of FIRMS experiments. (A) The mRNA carrying a translating ribosome is immobilized to the surface of a streptavidin-coated sample well via its 5′-biotin (blue). The nucleotides downstream from the mRNA entry site into the ribosome are accessible to hybridization to a DNA probe, whereas the nucleotides upstream inside the ribosome are not accessible. (B) Addition of a DNA probe to the immobilized mRNA allows formation of an mRNA-DNA duplex. The DNA probe is conjugated with a magnetic bead, allowing for magnetization of the ribosome complex by a permanent magnet, and showing magnetic ordering of the bead by the black arrow pointing up. The magnetic signal of the ribosome complex is measured by an atomic magnetometer. Because the magnetic bead is immobilized to the surface via the mRNA-DNA duplex, its binding strength depends on the number of base pairs of the duplex. Prior to the force-induced dissociation of the duplex, the ribosome complex gives a high strong magnetic signal. (C) When the applied centrifugal force exceeds the dissociation force of the mRNA-DNA duplex, the DNA probe is dissociated from the duplex, leaving its magnetic bead randomly orientated due to Brownian motion as indicated by the white arrow in the bead. A complete dissociation of the duplex gives a zero magnetic signal. Because the dissociation force of the mRNA-DNA duplex is proportional to the number of base pairs, the position of mRNA is revealed by the dissociation force as indicated by a decrease of the magnetic signal. Single base-pair resolution is routinely achieved by FIRMS to distinguish different reading frames (22). (D) A representative force spectrum that illustrates the high magnetic signal prior to duplex dissociation and the low magnetic signal after dissociation. The value of the dissociation force reveals the ribosome position on the mRNA, hence the reading frame.