Table 4.
EMD from predicted haplotypes to the true haplotype population and haplotyping method improvement
| (a) | |||||||
|---|---|---|---|---|---|---|---|
| Consensus | CliqueSNV | aBayesQR | PredictHaplo | ||||
| Benchmark | EMD | EMD | Impr. | EMD | Impr. | EMD | Impr. |
| HIV9exp | 4.18 | 2.35 | 1.78 | 5.02 | 0.83 | 6.90 | 0.61 |
| HIV2exp | 5.50 | 1.87 | 2.94 | 3.02 | 1.82 | 3.65 | 1.51 |
| HIV5exp | 14.80 | 7.37 | 2.01 | 14.05 | 1.05 | 9.43 | 1.57 |
| HIV7sim | 9.63 | 0.76 | 12.72 | 0.67 | 14.4 | 2.00 | 4.80 |
| IAV10sim | 4.22 | 0.59 | 7.2 | 3.57 | 1.18 | 2.97 | 1.42 |
| (b) | |||||||
| Consensus | CliqueSNV | 2SNV | PredictHaplo | ||||
| Benchmark | EMD | EMD | Impr. | EMD | Impr. | EMD | Impr. |
| IAV10exp | 4.22 | 0.22 | 19.18 | 0.23 | 18.35 | 0.38 | 11.12 |
Four haplotyping methods(aBayesQR, CliqueSNV, Consensus, PredictHaplo) are benchmarked using five MiSeq (A) and one PacBio datasets (B). The column Impr. (improvement) shows how much better is prediction of haplotyping method over inferred consensus, and it is calculated as , 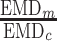 where EMDc is an EMD for consensus, and EMDm is an EMD for method.
where EMDc is an EMD for consensus, and EMDm is an EMD for method.
