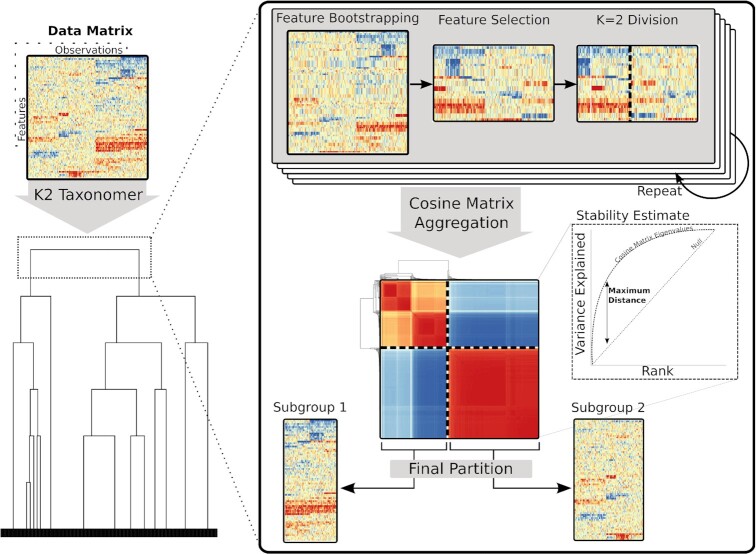
Figure 1.
Schematic of the K2Taxonomer recursive partitioning algorithm. For each partition, K2Taxonomer generates an ensemble of K = 2 estimates from the feature bootstrapped data followed by variability-based feature selection. This ensemble is aggregated to a cosine matrix followed by hierarchical clustering and tree cutting. A stability estimate, indicative of the consistency of K = 2 estimates, is calculated based on an eigendecomposition of the cosine matrix. See supplementary methods for a more thorough description of the elements of this procedure.