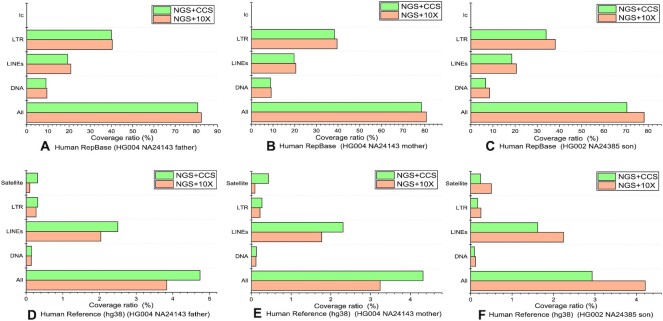
Figure 7.
Comparison between the detection results generated from the de novo mode of LongRepMarker based on three groups of NGS short reads + barcode linked reads (HG004_NA24143_father, HG004_NA24143_mother and HG002_NA24385_son) and the detection results generated from the de novo mode of LongRepMarker based on three groups of NGS short reads + SMS long reads (CCS) in terms of the proportion of covering the human RepBase library and the repetitive regions on the reference genome of human. The label All represents the total coverage ratio, which is the sum of the proportion of detection results covering all kinds of repetitive sequences in the corresponding library. The label DNA indicates the proportion of the detection results covering the DNA transposon elements-type repetitive sequences in the corresponding library, label LINEs indicates the proportion of the detection results covering the LINEs-type repetitive sequences in the corresponding library, label LTR indicates the proportion of the detection results covering the LTR-type repetitive sequences in the corresponding library, label lc indicates the proportion of the detection results covering the low complexity-type repetitive sequences in the corresponding library, and label Satellite indicates the proportion of the detection results covering the Satellite-type repetitive sequences in the corresponding library. Sub-figures (A) to (C) show the comparison of the ratio of the detection results of these two models on the three groups of hybrid sequencing data covering the human RepBase library. Sub-figures (D) to (F) show the comparison of the ratio of the detection results of these two models on the three groups of hybrid sequencing data covering the repetitive sequences on the reference genome of human.