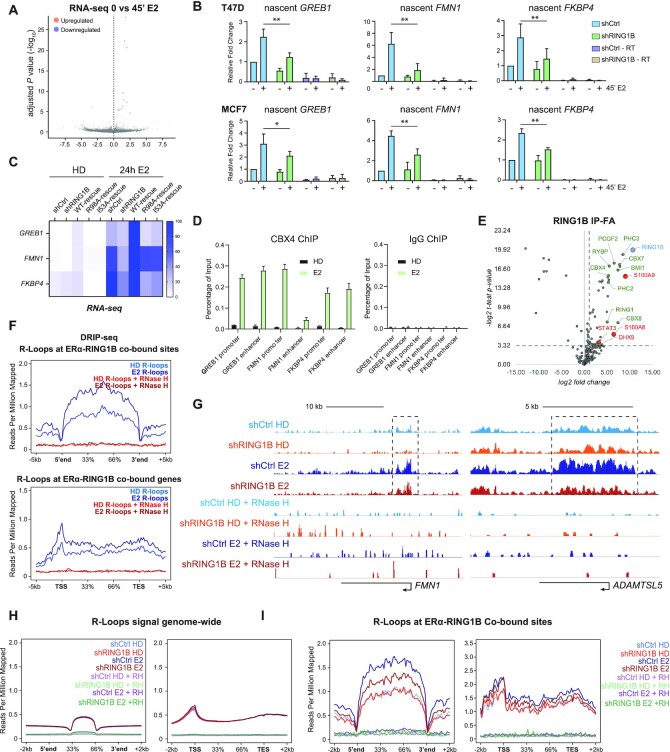
Figure 2.
RING1B promotes R-loop formation at ERα target genes via direct participation in their transcription. (A) RNA-seq volcano plot depicting one significantly downregulated and 11 significantly upregulated genes (P-value < 0.05, FC > 1.5) in parental T47D cells before and after 4′ of E2 stimulation; n = 2. (B) RT-qPCR measuring levels of GREB1, FMN1 and FKBP4 nascent transcripts in T47D and MCF7 shCTR and shRING1B cells before and after 45′ of E2 stimulation. shCTR – RT and shRING1B – RT are RNA samples subjected to the cDNA conversion process in the absence of reverse transcriptase. Enrichment detected in these samples represent genomic DNA contamination. Error bars represent the standard deviation of three biological replicates. *P-value < 0.05, **P-value < 0.01, one-tailed paired t-test. (C) RNA-seq heatmap depicting expression of GREB1, FMN1, and FKBP4 in T47D shCTR, shRING1B, as well as shRING1B cells rescued with wildtype (WT), nucleosome-binding mutant (R98A) and a catalytic-dead mutant (I53A) RING1B; n = 2. (D) CBX4 and IgG, as a negative control, ChIP-qPCR of RING1B/ERα co-bound sites before and after 45 min of E2 administration; n = 2. (E) Endogenous RING1B immunoprecipitation with nuclear extracts after crosslinking with 1% FA for 10 min. Proteins bound to RING1B were identified by LC-MS/MS, and enrichment was calculated based on fold change over IgG enrichment and P-value < 0.05. IgG was used as a negative control. Experiments were performed in three biological replicates. Proteins labeled in green are PRC1 subunits. Proteins labeled in red are new RING1B-associated proteins. (F) Average DRIP-seq signals in parental T47D cells before and after 45′ of E2 stimulation at ERα and RING1B co-bound sites (top) and genes (bottom). No signal was detected in samples treated with RNase H, indicating that the signal observed was specific for R-loops. (G) T47D shCTR and shRING1B DRIP-seq signal before and after 45 min of E2 at the FMN1 and the ADAMTSL5 genes. (H) Average genome-wide DRIP-seq signal (left) and with respect to genes that contain R-loops (right). (I) Average DRIP-seq signal at all RING1B and ERα co-bound sites (left) and genes (right) that contain R-loops.