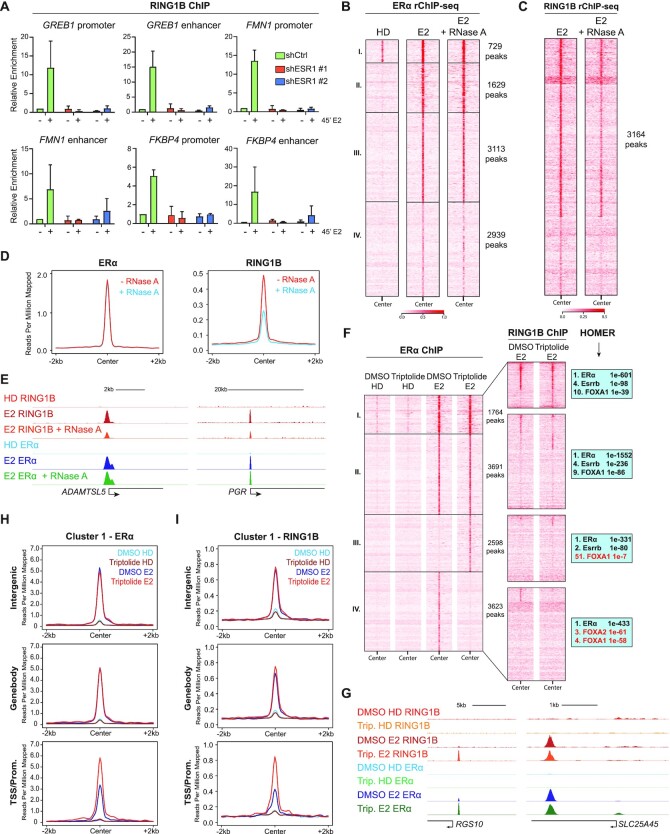
Figure 4.
RING1B recruitment to chromatin is dependent on ERα and chromatin-associated RNA. (A) RING1B ChIP-qPCR in shCTR and 2 shESR1 T47D cell lines before and after 45 min of E2 administration at six sites with known RING1B and ERα co-occupancy. (B and C) rChIP-seq heatmap depicting E2-induced ERα (B) and RING1B (C) chromatin binding at the previously described 4 clusters (Figure 3A); n = 2. (D) Average signal of ERα and RING1B chromatin binding after 45 min of E2 stimulation with or without RNase A treatment. (E) RING1B and ERα ChIP-seq signal after 45 min of E2 stimulation with or without RNase A treatment at ADAMTSL5 and PGR. (F) ERα (left) and RING1B (center) ChIP-seq heatmaps before and after 45 min of E2 treated with vehicle (DMSO) or 10 μM triptolide for 9 h clustered by ERα occupancy. HOMER motif analysis of the four clusters (right). Cluster III ERα sites contain very little FOXA1 motif enrichment. (G) RING1B and ERα ChIP-seq before and after 45 min of E2 in cells treated with DMSO or triptolide showing increased ERα and RING1B recruitment at the RGS10 promoter but little change at the SLC25A45 gene body. (H–I) Average signal of ERα (H) and RING1B (I) at cluster I, classified by genomic annotation, before and after 45 min of E2 in cells treated with DMSO or triptolide.