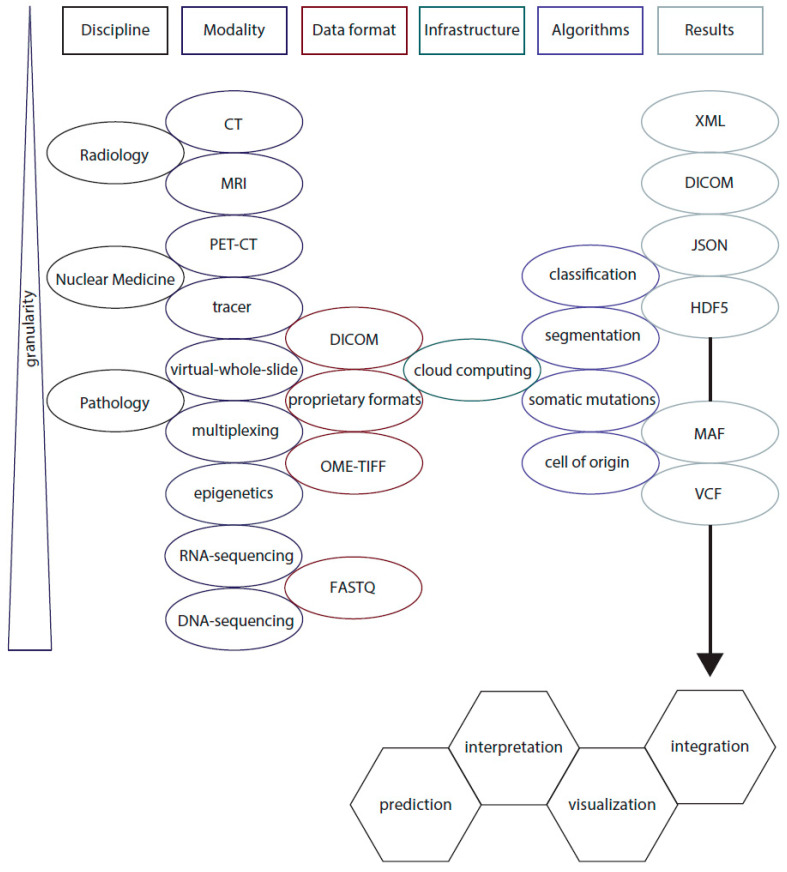
Figure 1.
Schematic of integrative data analysis in oncology. Diagnostic medical disciplines, including Radiology, Nuclear Medicine and Pathology are gathering data using their different modalities, like computer tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET-CT), as well as tracers that are able to highlight molecules of interest. While Radiology and Nuclear Medicine share a sophisticated data format of digital imaging and communications in medicine (DICOM) objects, which includes relevant (pseudonymized) patient (meta) data, other formats from virtual-whole-slide images and multiplexing techniques may be stored using proprietary vendor formats. The maturing file format of open microscopy environment tagged file format (OME-TIFF) appears to be especially interesting as future solution, as DICOM objects for virtual-microscopy-images have not yet been broadly accepted by the community [97,112,114]. Cloud computing services would require specialized structures, like central processing units (CPU) and graphical processing units (GPU) that meet the desired calculating capabilities. In the near future, several specialized algorithms will be executed allowing segmentation of regions of interest and classification of image objects, in addition to accurate variant calling and methylome-based cell of origin determination using epigenetics. The given results of image analysis are stored using extensible markup language (XML), DICOM objects or JavaScript Object Notation (JSON), together with Hierarchical Data Format (HDF5). Here, spatial descriptive statistics of cellular and subcellular structures of interest are calculated. Data formats like variant calling files (VCF) and Mutation Annotation Format (MAF; https://docs.gdc.cancer.gov/Data/File_Formats/MAF_Format/, accessed on 3 September 2021) may be used to store results of DNA-sequencing. Finally, the data need to be integrated and visualized [115,116] to allow interpretation in order to allow patient stratification, identifying risk groups of cancer patients and to predict therapy responses. Given the increasing complexity within oncology, analytical tools are needed to retrieve current information on drug targets, clinical trials and potential treatment options, which may need to be integrated in a given workflow [117,118].