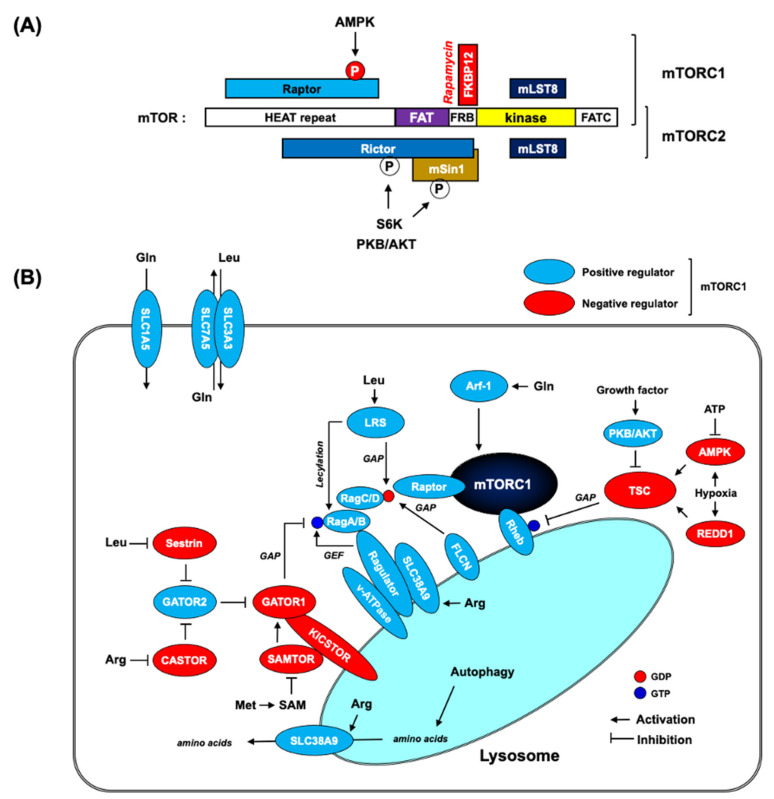
Figure 2.
mTORC and its nutrient signaling modules. (A) A schematic representation of mTOR domain structure and mTORC subunits. mTOR is composed of a huntingtin-elongation factor 3-regulatory subunit A of PP2A-TOR1 (HEAT) repeat, a FRAP-ATM-TRRAP (FAT) domain, a FKBP12-rapamycin binding (FRB) domain, a catalytic domain (kinase), and a FAT domain at the C terminus (FATC). mTORC1 includes mTOR, regulatory-associated protein of mammalian target of rapamycin (Raptor), and mammalian lethal with sec-13 protein 8 (mLST8). Raptor is a phosphorylation target of AMPK, which leads to mTORC1 inactivation upon AMPK activation. mTORC2 contains mTOR, rapamycin-insensitive companion of mTOR (Rictor), mammalian stress-activated MAP kinase-interacting protein 1 (mSin1), and mLST8. The binding of Rictor and mSin1 masks the FRB domain on mTOR to prevent FKBP12–rapamycin binding, thereby rendering mTORC2 insensitive to rapamycin. mTORC2 is shown to be phosphorylated by PKB/AKT, as well as a downstream kinase S6K in mTORC1 signaling, but it is debated whether their physiological significance entails activation or inhibition. (B) mTORC1 and amino acid signaling network. mTORC1 is activated by Rheb on lysosomes. Therefore, localization of mTORC1 is required for activation, which is dependent on the intracellular amino acid level. Rag GTPase is a major arm for transmitting information on intracellular amino acid into mTORC1. In an amino acid-rich condition, the active Rag GTPase (GTP-loaded RagA/B and GDP-loaded RagC/D) binds to Raptor to recruit mTORC1 onto lysosomes. Many different amino acid-sensing molecules directly or indirectly regulate GTP/GDP loading status on Rag GTPase complex. Additionally, Rheb is negatively regulated by the TSC complex, a GAP for Rheb. The TSC complex integrates many different inputs (growth factors, cellular energy level, and oxygen level) to fine-tune Rheb for the tight regulation of mTORC1 signaling in response to various extracellular and intracellular cues. The proteins shown in the figure are not drawn to scale.