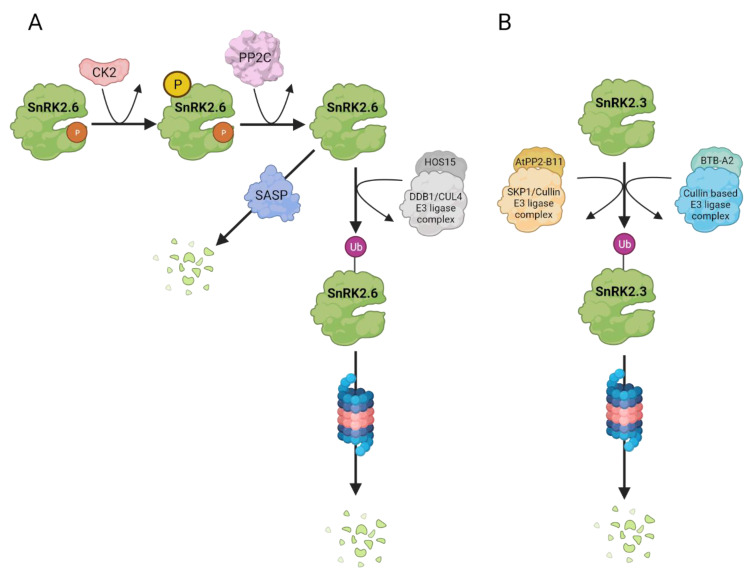
Figure 4.
Controlled degradation of SnRK2s in Arabidopsis. ABA-responsive SnRK2s undergo degradation in a kinase-specific manner via proteasomal degradation pathway as well as by specific proteases. (A) SnRK2.6 ubiquitination occurs when the kinase is in an inactive, dephosphorylated state. The kinase interacts with HIGH OSMOTIC STRESS 15 (HOS15), a CULLIN4 (CUL4)-DAMAGED DNA BINDING PROTEIN1 (DDB1)-based E3 ubiquitin ligase and is targeted to degradation via Ubiquitin Proteasome System (UPS). Phosphorylation of SnRK2.6 within the ABA-box, catalysed by Casein Kinase 2 (CK2), promotes SnRK2.6 degradation by enhancing its interaction with and dephosphorylation by clade A PP2Cs, and thus interaction of SnRK2.6 with HOS15. Additionally, SnRK2.6 protein level is regulated via Senescence-Associated Subtilisin Protease (SASP). (B) SnRK2.3 stability is negatively controlled by PHLOEM PROTEIN 2-B11 (AtPP2-B11), an F-box component of the SKP1 (S PHASE KINASE-ASSOCIATED PROTEIN 1)/Cullin/F-box E3 ubiquitin ligase complex as well as Broad-complex, Tramtrack, and Bric-a-brac proteins, namely BTB-A2.1, BTBA2.2, and BTB-A2.3, substrate adaptors for cullin-based E3-ligases. The ubiquitinated kinase is degradated by UPS.