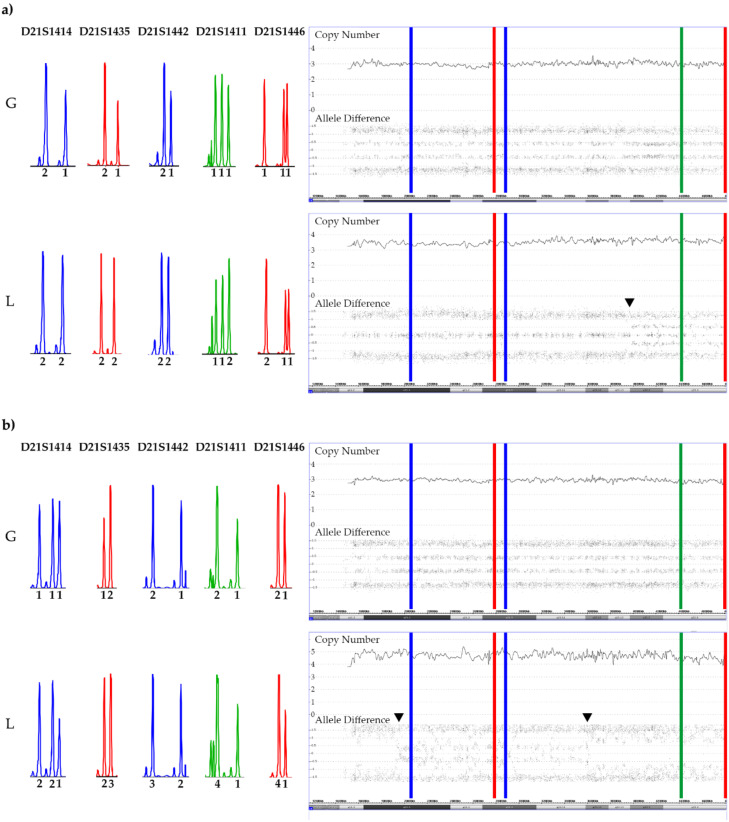
Figure 10.
SNP array and STR patterns of the leukemic and remission sample of constitutional trisomic patients with an acquired tetrasomy (a) or pentasomy (b) of chromosome 21. The corresponding allele copy numbers are indicated below the respective STR markers. As evidenced by the STR markers with three alleles, both cases result from a (most likely maternal) meiosis 1 error. However, without parental material we were unfortunately not able to unequivocally define which parental homologues were indeed gained in the tetra- and pentasomic clones, respectively. In the tetrasomic case (a), the first four STR markers on the right and the fifth one on the left define the duplicated chromosome. Moreover, in this example a meiosis 1-associated switch from a “2 + 2” to a “2 + 1 + 1” configuration uncovered a recombination site that is clearly visible in the array-derived allele distribution pattern, and which is incidentally located in the ERG gene (indicated by the black arrow). Copy number validation with a RUNX1 FISH probe identified a trisomy in 21% and a tetrasomy in 79% of the leukemic cells. The situation is similar in the pentasomic case (b), in which two of the four STR markers with homozygous alleles (“2 + 1”; D21S1435, D21S1442, D21S1411and D21S1446), which might have been either uniparental or biparental-derived, became duplicated (D21S1411and D21S1446) together with two of the three D21S1414 and one each of the D21S1435, D21S1442 markers. There are two meiotic recombination sites, which in this instance resulted from a “4 + 1” to a”2 + 3” and again a “4 + 1” allele distribution switch (indicated by the black arrows). Copy number validation with a RUNX1 FISH probe identified a trisomy in 29% and a difficult to discriminate tetra- or pentasomy in 71% of the leukemic cells. We found such recombination sites in two of the three tetrasomic and in two of the four pentasomic cases.