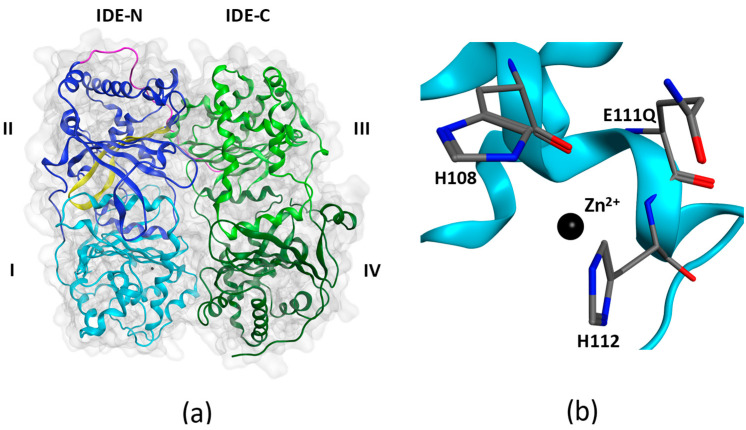
Figure 1.
X-ray structure of insulin-degrading enzyme in its closed conformational state (PDB: 4PES). (a) IDE is composed of an N-terminal half (IDE-N) containing domains I and II, and a C-terminal half (IDE-C) containing domains III and IV. The two halves are joined by an unstructured linker (magenta). Domain I contains the HXXEH zinc-binding and catalytic motif. Domain II contains the exosite (yellow) in an anti-parallel β-sheet conformation. (b) The HXXEH motif includes H108 and H112 that coordinate Zn2+ and the catalytically important glutamate (E111) residue. When E111 is mutated to a glutamine as in 4PES, the enzyme becomes inactive.