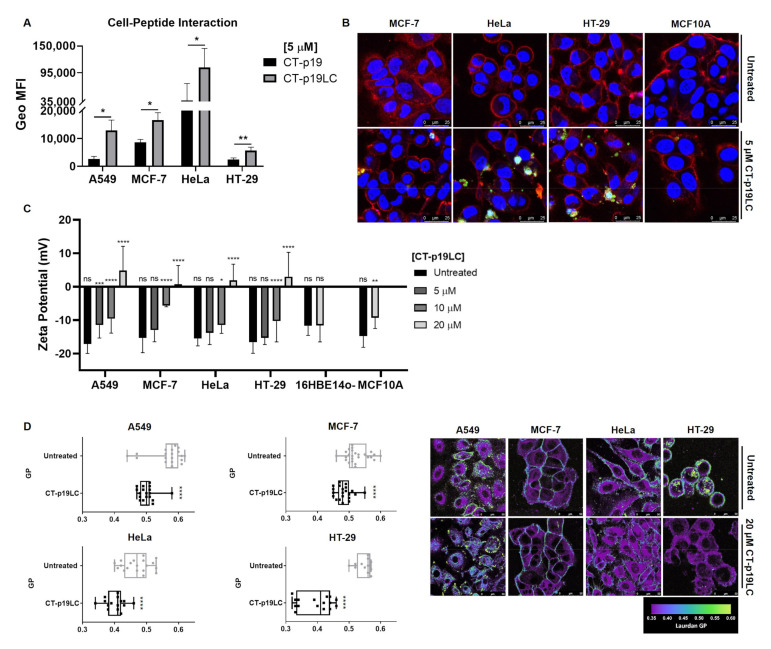
Figure 4.
CT-p19LC membrane-active properties: (A) flow cytometry quantitative analysis of cancer cell–peptides interaction. Results are reported as the means ± SD, and each condition had at least n = 3. * and ** denote significant differences of p < 0.1 and p < 0.01, respectively, when comparing CT-p19LC treatment with CT-p19 treatment; (B) representative confocal microscopy qualitative analysis of CT-p19LC cellular uptake by MCF-7, HeLa and HT-29 cancer cells and MCF10A non-cancer cells incubated with PBS pH 7.4 as control and 5 μM of peptide labeled with 5,6-FAM (green color) for 2 h. WGA Alexa Fluor® 633 and Hoechst 33342, for staining the plasma membrane and nucleus, respectively, are shown in red and blue colors. Scale bars represent 25 μm; (C) zeta potential of A549, MCF-7, HeLa and HT-29 cancer cells and 16HBE14o- and MCF10A non-cancer cells in the presence of CT-p19LC peptide. A total of 1.5 × 105 cells/mL were incubated and stabilized for 30 min at 37 °C with different peptide concentrations, and the zeta potential was measured. Data are represented as means ± SD. *, **, ***, **** and ns denote significant differences of p < 0.1, p < 0.01, p < 0.001 and p < 0.0001 and differences that were not statistically significant, respectively, when comparing the untreated condition (0 μM) with increasing concentrations of CT-p19LC (5, 10 and 20 μM); (D) the effects of CT-p19LC on the cell‘s membrane order for A549, MCF-7, HT-29 and HeLa cancer cell lines and their respective GP values. All represented cell lines were seeded on μ-Slide 8-well glass-bottom chambers and treated with 20 μM of CT-p19LC for 2 h. For each condition, 5 μM of Laurdan was used. Untreated cells were used as the control. Homemade software built in a MATLAB environment was used to measure the GP values. Representative Laurdan GP images are shown. Scale bars represent 50 μm. Average GP values are expressed as means ± SD from at least 15 individual cells in each condition. Results are compared to the untreated population with equal variance (****, p < 0.0001).