Table 2.
The results of molecular docking between the active site of BACE1 (PDB ID: 2WJO) and candidate ligands.
| Ligand | Binding Energy (kcal/mol) | Inhibition Constant | Amino Acid Interaction | ||
|---|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Bond | Electrostatic Bond | |||
Quercetin (reference inhibitor)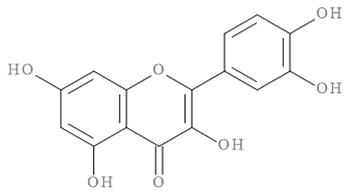
|
−8.78 | 365.72 µM | VAL31 GLN73 (2) PHE108 (2) |
LEU30 (2) TRP115 |
- |
3-O-Methylgallate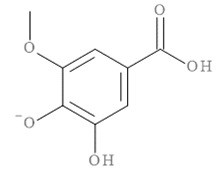
|
−5.20 | 154.27 µM | THR72 GLN73 (3) PHE108 THR232 (2) |
PHE108 | - |
4-Aminomethylindole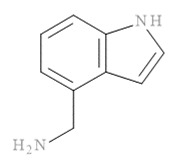
|
−6.49 | 17.63 µM | - | TYR71 (2) GLY230 THR231 |
- |
Bergenin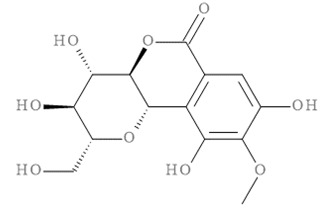
|
−8.52 | 572.8 nM | TYR71 THR72 GLN73 THR232 |
- | - |
Clausarinol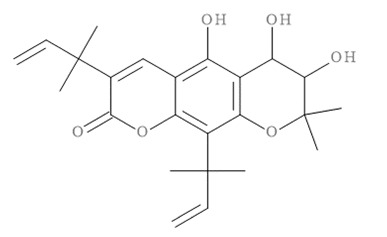
|
−9.96 | 50.27 nM | THR72 (2) GLN73 (2) THR232 (2) ASN233 |
LEU30 PHE108 (2) ILE118 ARG235 |
- |
Emmotin A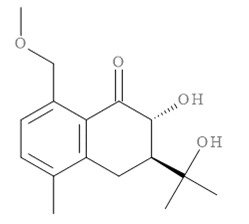
|
−7.90 | 1.62 µM | THR72 (3) THR232 THR231 |
TYR71 PHE108 ILE118 |
- |
Gallic acid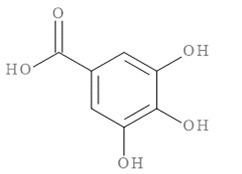
|
−5.09 | 186.3 µM | THR72 GLN73 (2) THR232 (2) |
- | - |
N-D-Glucosylarylamine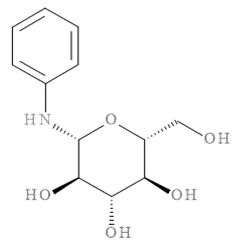
|
−7.10 | 6.29 µM | GLN73 (2) PHE108 GLY230 THR231 (2) THR232 (2) |
- | ASP228 |
Quercetin-3′-glucuronide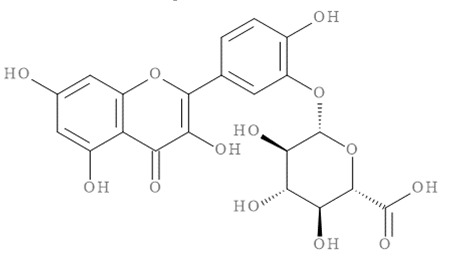
|
−10.74 | 13.45 nM | TYR71 THR72 GLN73 (3) ASP228 ARG235 (2) |
LEU30 (2) ALA335 |
- |
Quercetin-3-O-glucoside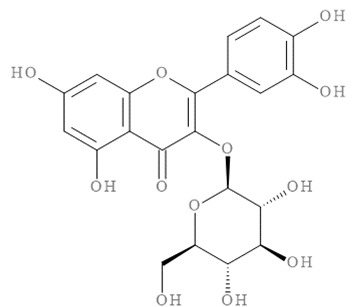
|
−10.43 | 22.54 nM | GLY34 GLN73 PHE108 (2) THR232 (3) ARG235 |
ASP228 | TYR71 |
Theogallin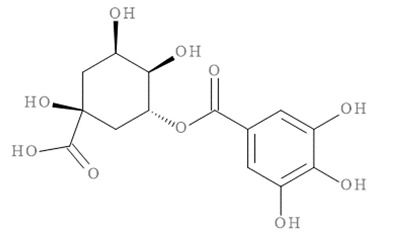
|
−8.97 | 266.1 nM | THR72 (3) GLN73 (3) ASP32 SER35 LYS107 PHE108 (2) THR231 |
TYR71 PHE108 |
- |
