Fig. 6 |. Applications for labelling different molecule types in cells.
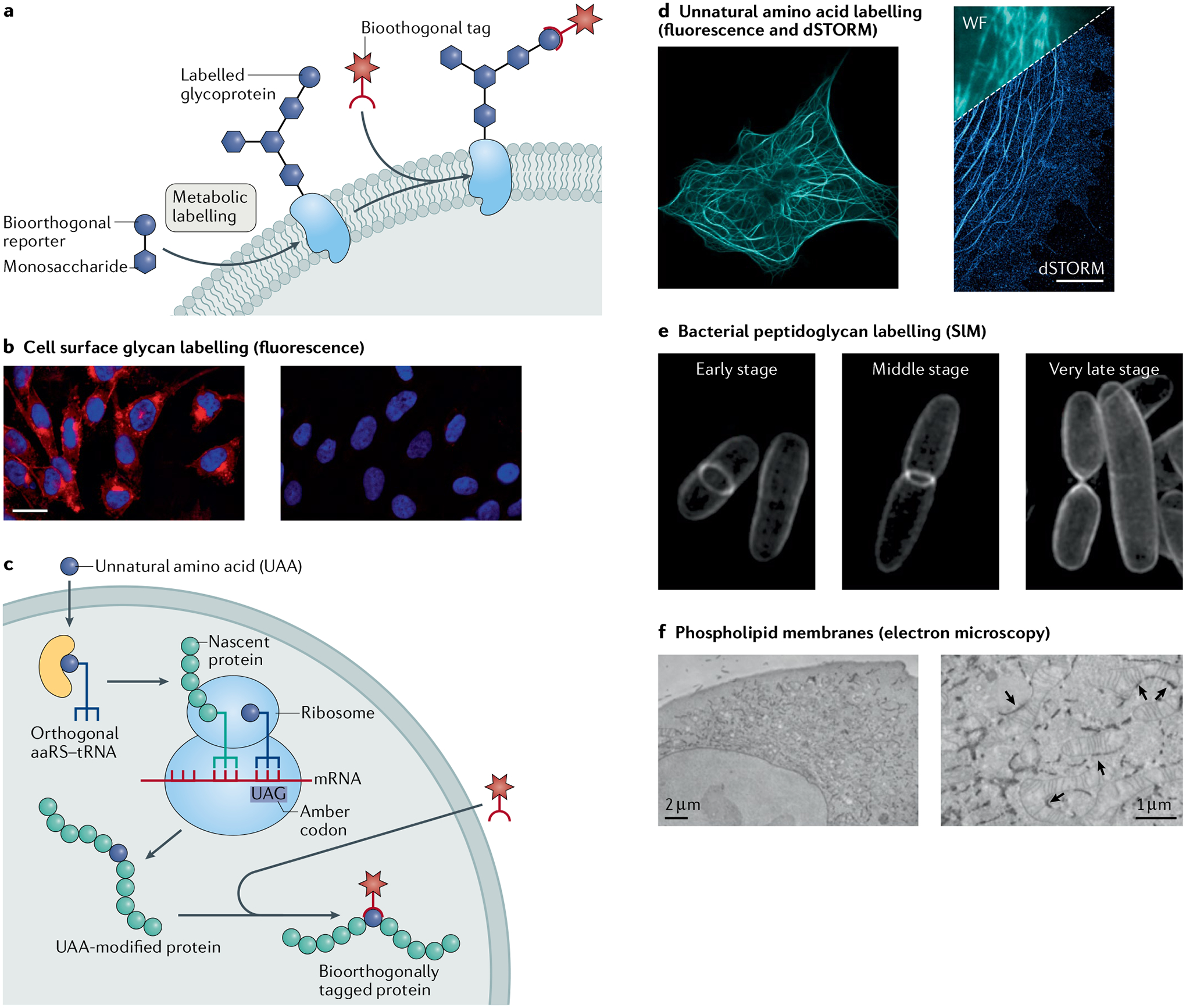
a | Model for metabolic engineering for cell labelling and imaging. b | Fluorescence microscopy of CHO cells incubated in the presence (left) or absence (right) of peracetylated N-azidoacetylmannosamine (Ac4ManNAz) and labelled with a fluorophore by the Staudinger ligation. c | Model for genetic code expansion as a strategy for cell labelling and imaging. d | Fluorescence and direct stochastic optical reconstruction microscopy (dSTORM) super-resolution images of COS-7 cells where microtubule-associated protein was encoded with an unnatural trans-cyclooctene (TCO) amino acid and tetrazine ligation was used to attach a microscopy dye. e | Structured illumination microscopy (SIM) images of Escherichia coli, where N-azidoacetyl-muramic acid (NAM) was metabolically incorporated into the bacterial peptidoglycan and fluorophore-labelled by copper(I)-catalysed azide–alkyne cycloaddition (CuAAC). f | Electron microscopy images of HeLa cells, where azido-choline was metabolically incorporated, and cyclooctyne/azide click chemistry was used to conjugate electron microscopy imaging agents. The arrows indicate sites of endoplasmic reticulum–mitochondria contacts. aaRS, aminoacyl-tRNA synthetase; WF, widefield image. Images in panel b adapted with permission from REF.252, ACS. Images in panel d reprinted from REF.176, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Images in panel e reprinted from REF.286, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Images in panel f reprinted from REF.296, Springer Nature Limited.
