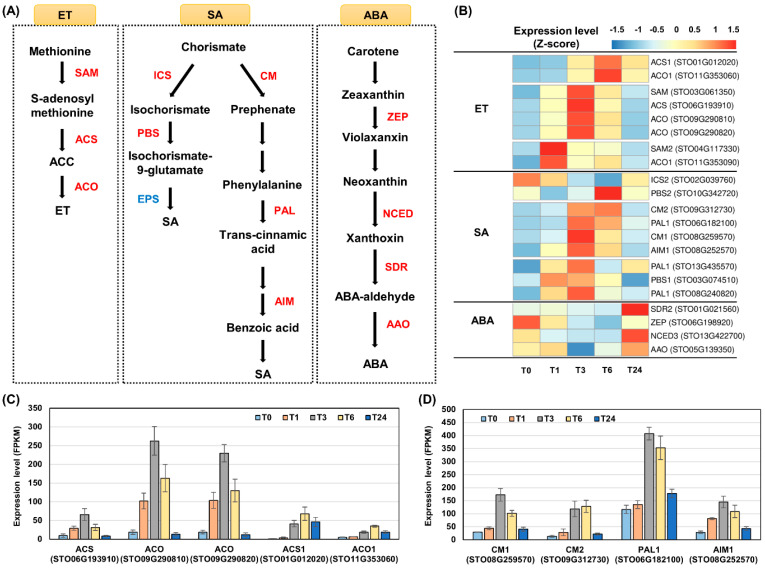
Figure 4.
Expression analysis of genes involved in ethylene (ET), salicylic acid (SA), and abscisic acid (ABA) biosynthesis induced by wounding in S. tora leaves. (A) ET, SA, and ABA biosynthesis pathways in plants. (B) Heatmap analysis of genes involved in ET, SA, and ABA biosynthesis. ACC, 1-aminocyclopropane-1-carboxylate; SAM, S-adenosylmethionine synthase; ACS, 1-aminocyclopropane-1-carboxylate synthase; ACO, 1-aminocyclopropane-1-carboxylate oxidase; ICS, isochorismate synthase; PBS, PphB SUSCEPTIBLE; EPS, ENHANCED PSEUDOMONAS SUSCEPTIBILITY; CM, chorismate mutase; PAL, phenylalanine ammonia-lyase; AIM, fatty acid beta-oxidation multifunctional protein; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; SDR, short-chain dehydrogenase/reductase; AAO, ABA aldehyde oxidase. The gene encoding the SA biosynthesis enzyme EPS (blue color) was not identified in this study. The fragments per kilobase of transcript per million mapped reads (FPKM) values of the individual genes were normalized to the Z-score, and the heatmap was visualized using the pheatmap package in R. (C) Expression of the ET biosynthesis genes. (D) Bar graph analysis of genes involved in second SA biosynthesis pathway. Statistical analysis was performed on the expression levels of each gene expressed at the different times compared with T0 (Table S2).