Figure 2.
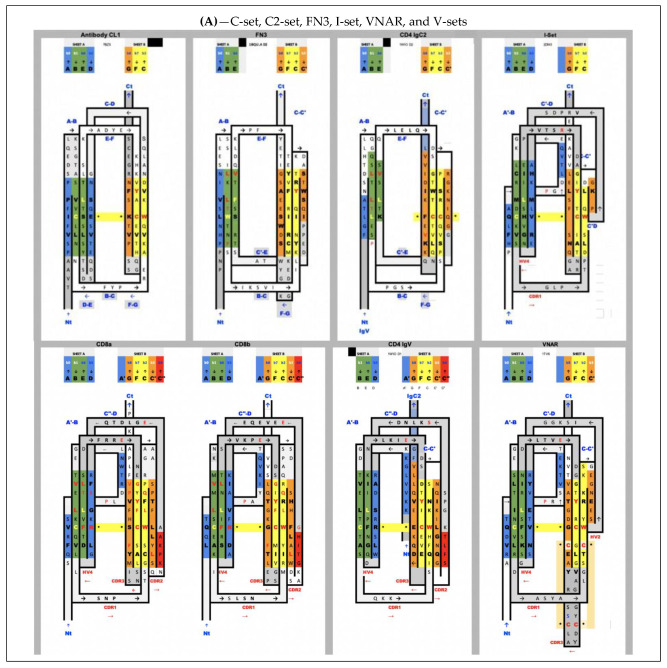
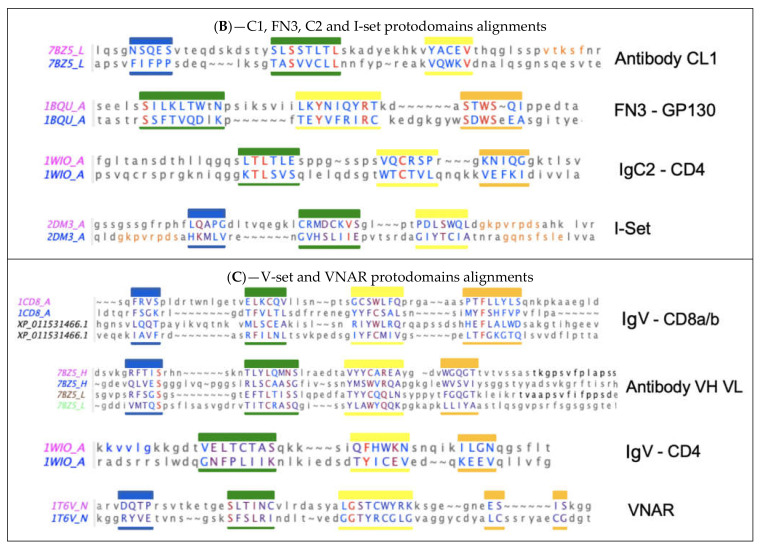
Sequence/topology maps comparison of topological variants of single Ig domains. (A) Sequence/topology maps: C1-set, C2-set, FN3, I-set, VNAR, and V-set for CD8a/CD8b and for CD4. See text for details on differences. Capital letters are used for structurally aligned residues. Colors are the same for matching strands in protodomains: blue for strands A and D, green for B and E (a full sheet ABED will read blue–green–green–blue), yellow for strands C and F, orange for strand C’ and G, and red for C” “IgV “linkers”. Colors green and yellow of central strands highlight the self-assembly of strand B-E and C-F of protodomains. FN3 and IgC2 have a similar topology, but FN3 does not share the CCW(L) present in all the selected Ig domains highlighted in central strands. Some IgV domains, such as CD2, however, can also miss that pattern (not shown). IgVs have a C” strand highlighted in red. We have highlighted CD8a and CD8b that form either homo- or heterodimers, with a split A/A’ strand where A’ swaps from one sheet, i.e., antiparallel to strand B and parallel to strand G, while CD4 IgV represents another type where the strand A is only on one sheet in parallel to strand G. VNAR shares the split strand A/A’ of IgVs, which is also present in the Ig-I set. The I-set is very similar to VNARs that have only a few more residues in their HV2 region that encompass a short C’ strand antiparallel to C, and a C’-D linker that is structurally superimposable in VNAR and I-set. In fact, the overall structural alignment is very good with an RMSD of 1.83A and a surprisingly good sequence match (https://structure.ncbi.nlm.nih.gov/icn3d/share.html?c8F4iowua2TAD6ej8, accessed on 27 August 2021) (B) Alignment of protodomains in C1-, C2- and I-set domains. Colors, as in A, show the corresponding strands aligning 3 out of 4 strands due to symmetry breaking (see text). In the I-set, the G strand is shown in orange, as well as that which would be C’ when comparing with IgVs, if it were not a protodomain “linker” in the I-set context. RMSD for protodomain alignments are for C1: 1.19 A, FN3: 1.8 A, C2: 1.98 A, I: 2.46 A (iCn3d self-alignment of 2DM3: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?tBzknU8spazpwCup7, accessed on 27 August 2021) (C) Alignment of protodomains in IgVs and VNAR domains. The symmetry breaking (see text) also extends to CD4-like IgVs, where strand A is swapped in protodomain 1 vs. strand D in protodomain 2. In other IgVs, such as CD8 or antibody VH-VL domains that we added for completeness, or in VNAR, the strand A is split A/A’, as in the I-set, and keeps a symmetric correspondence with strand D, overall, in all 4 strands AB-CC’ vs. DE-FG, with only a partial symmetry breaking through strand A’. Protodomain alignments consistently show a structural match for conserved and co-evolved residues C in strand B and strand F vs. residues W and (most often) L in strand C and strand E, respectively. RMSD for alignments: for CD8a-IgVs, 1.61 A (CD8b sequences are indicated sequence by homology with mouse CD8b structure (2ATP) whose protodomains superimpose with and RMSD of 0.895 A and 1.54 A), followed by VH-VL protodomains for reference with an RMSD of 1.8, 0.882, and 1.74 A respectively; CD4-IgV protodomains, 1.84 A; VNAR protodomains, 2.31 A.