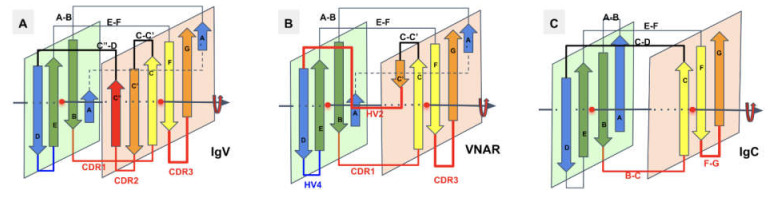
Figure 4.
Ig domain topologies for IgV, Shark VNAR, and IgC. (A) In IgV domains, the A strand has a flexible hinge in the middle, usually a cis-proline or a stretch of glycines and swaps the upper part of the strand (noted A’) to Sheet B. (B) VNAR shows that same domain level organization with two protodomains, yet a much smaller inter-protodomain linker, eliminating the linker’s secondary structure as present in IgV domains: the C” strand and the CDR2 loop. Instead, a short HV2 is observed. In the literature, the C’ strand is usually included in the HV2 region, as it is extremely short. In addition, a hydrophilic set of residues facing out on Sheet B (GF|CC’), rather than being hydrophobic in IgV, do not permit the formation of a symmetric dimer. Dimerization in crystal structures can be observed through HV2, yet they may not be relevant biologically. (C) IgC. Here, we consider only the IG C1 set, i.e., the antibody-constant domain-like set, to exemplify an additional protodomain connectivity. In this case, the final domain is formed by a full 4-stranded AB|ED (Sheet A) vs a 3-stranded GFC Sheet B, and no C’ strand. Interestingly, this enables the domain-level dimerization through that 4-stranded Sheet A to form an 8-stranded barrel, as opposed to the IgV that uses the opposite Sheet B to form an 8-stranded dimer barrel.