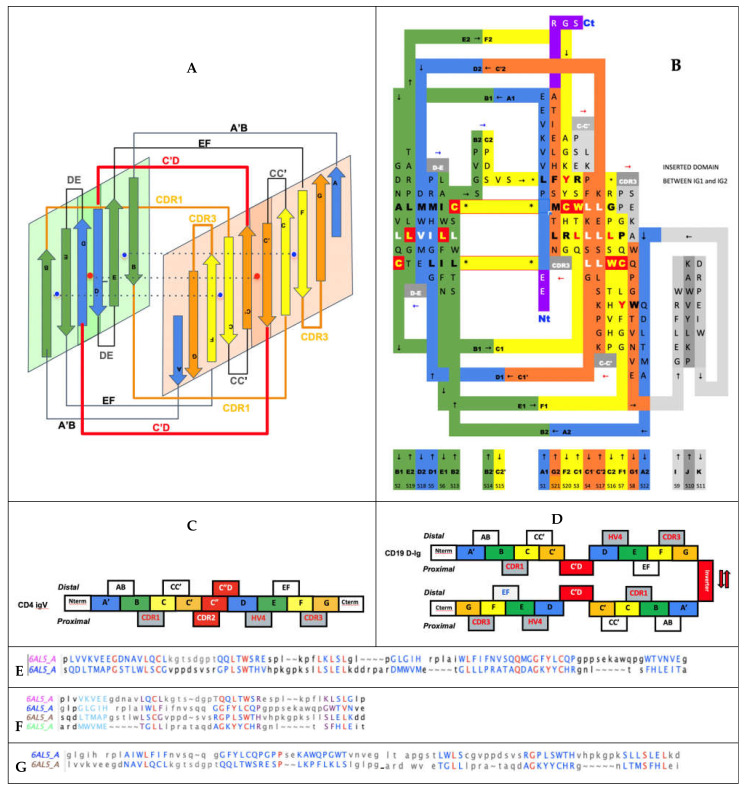
Figure 6.
Double IgV domain topology (CD19). (A) CD19 double Ig domain topology: Topology diagram with the two sheets facing each other. The central axis of symmetry (red dot) and the two local ones (blue dots) are shown in dashed lines. (B) The topology/sequence map showing the pairing of individual residues in complementary beta strands. The comparison with the topology/sequence map for a single Ig domain in Figure 1D shows a conserved organization of composite Ig domains within the double domain, including the CCWL pattern (highlighted in red), as it is in a swapped dimer in Figure 5C. (C) Sequence of an IgV with an A’ and no A strand as in CD4, marking the sequence stretches for strands and loops. The (CDR2-C”-C”D loop) linker inverts protodomain 2 A’GFCC’ vs protodomain 1 BED. The CDR loops CDR1, CDR2, and CDR3 and HV4 are all on the same side, here defined as proximal for comparison. (D) The CD19 sequence composed of protodomain 1 AB-CC’ and protodomain 2 DE-FG in parallel with a C’D short linker. In the first, a sequential parallel Ig domain CDR1 on the proximal side vs. CDR3 and HV4 on the distal side. The second duplicated parallel Ig domain composed of protodomain 3 and 4 is inverted through a small, inserted domain (in grey in (B)). This positions the two parallel Igs to intercalate and form a double Ig domain formed by two long sheets: (BED)1(DEB)2 vs. (A’|GFCC’)1(C’CFG|A’)2. In doing so, two fused composite Ig domains are formed combining p1 + p4 as an Ig domain of topology (AB-CC’|DE-FG) and p2 + p3 as an inverted Ig domain with the topology (DE-FG|AB-CC’), since, in sequence, p2 (DE-FG) precedes p3 (AB-CC). The variable regions (CDR1, CDR3, and HV4) for the first composite Ig domain (p1 + p4) are on the proximal side, as is the case for a regular IgV domain in (C), while in the second inverted Ig domain (p3 + p4), variable regions are on the distal side. (E) Structural alignment of the two parallel Ig domains within CD19, i.e., p1 + p2 vs. p3 + p4 matching within 1.65 A RMSD. (F) Alignment of the 4 individual Ig protodomains: RMSD p1 vs. p2: 1.99 A, p3: 1.29 A, p4: 1.60 A on the 3 last strands BCC’ vs. EFG (since A’ strands are not structurally matched to D strands—see symmetry breaking discussion; they are indicated in light blue). (G) Alignment of the two structural Ig domains within the double Ig (CD19). The first p1 + p3 matches the second inverted Ig p2 + p4 within 2.56A RMSD.