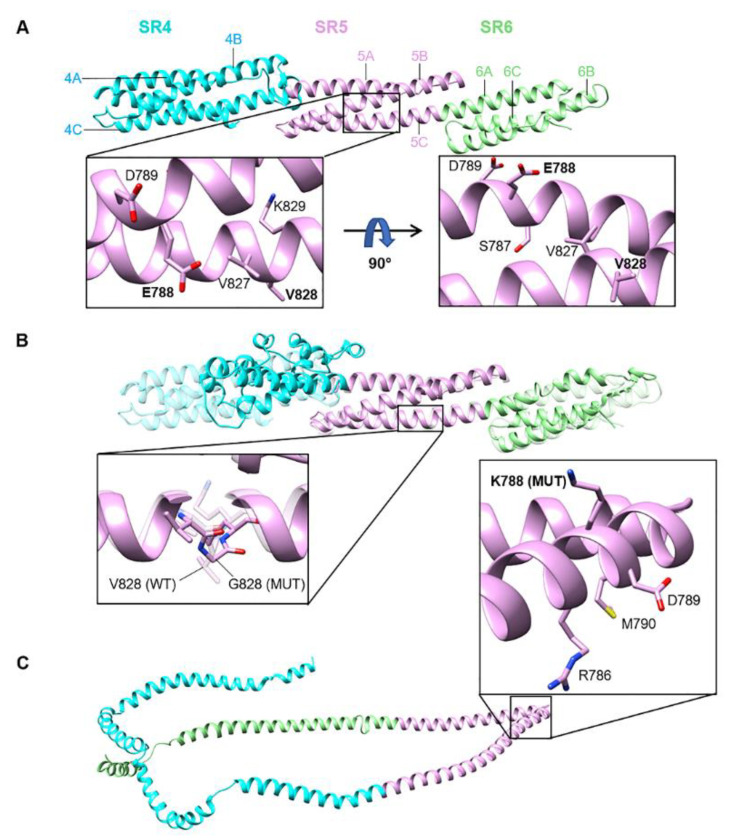
Figure 2.
Structural models of spectrin repeats SR4-SR5-SR6 for wild-type and mutant nesprin-2 giant proteins. (A) Wild-type (WT) SR4-SR5-SR6 with major helices labelled. WT residues within the affected region of SR5 (helices 5B and 5C) are presented in two orientations (90°-rotated; inset). Residues E788 and V828 (altered in mutants) are shown in bold. (B) Superposition of V828G mutant (solid) and WT (transparent) SR4-SR5-SR6 structures. SR4 and helix 6B (SR6) conformations are distorted in the V828G mutant, whilst SR5 is structurally homologous to WT. The peptide backbone (as well as residue side-chains) is presented in ‘stick’ form for positions 827–829 to distinguish between mutant glycine and WT valine at position 828 (inset). G828 mutant (MUT) and V828 (WT) residues (labelled) are depicted as solid and transparent structures, respectively. (C) E788K mutant SR4-SR5-SR6, showing complete loss of the typical triple-helical SR structure. K788 (MUT) and neighbouring residues are shown (inset). A superposition with the WT structure was not possible, due to extreme conformational differences. All structural models were predicted using PHYRE2. Stick representations are coloured by element (bound hydrogens are not depicted): carbon = pink, nitrogen = blue, sulfur = yellow, oxygen = red. For all structures, SR4 = turquoise, SR5 = pink, SR6 = green.