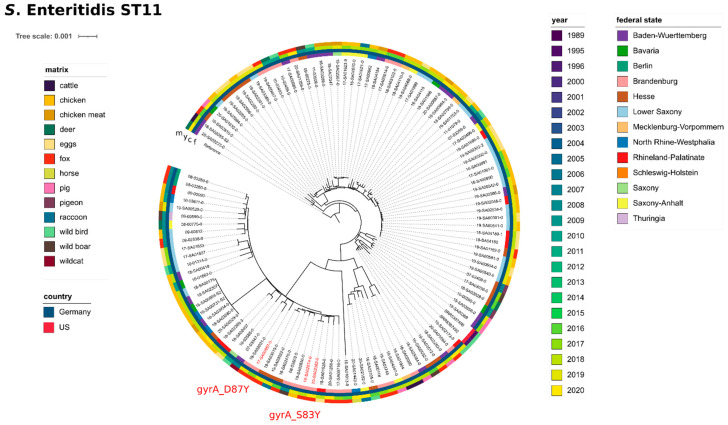
Figure 4.
Phylogenetic relationship between S. Enteritidis (ST11) wildlife isolates and isolates from food, feed and farm animals. Maximum likelihood phylogenetic tree based on core SNPs in the bacterial chromosome. The chromosome sequence of S. Enteritidis strain ATCC BAA-708 (NZ_CP025554.1) was used as reference. The tree was computed with the snippySnake pipeline with IQ-TREE, visualized in iTOL and manually rooted to the reference. The colour bars (inside to outside) identify the German federal state (f), national country (c), sampling year (y) and matrix (m) of the samples. Isolates carrying antimicrobial resistance genes are highlighted in colour with the type of resistance genes indicated. The scale bar shows the genomic distances of the sequences in substitutions per site.