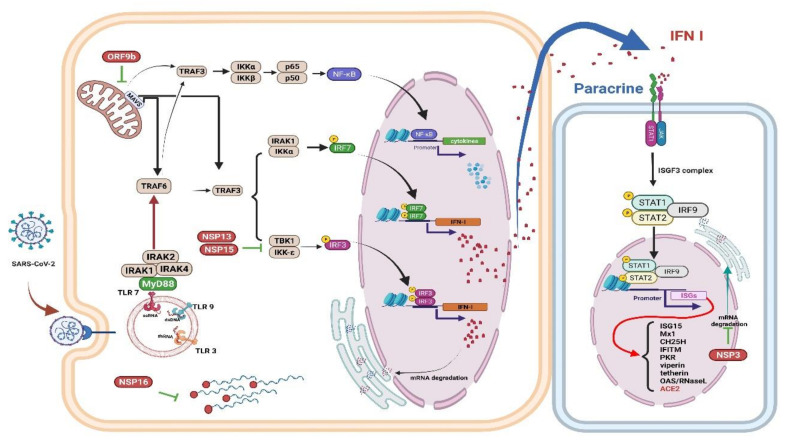
Figure 2.
Immune evasion by SARS-CoV-2. The schematic shows how SARS-CoV-2 viral proteins are able to inhibit various immune processes such as pathogen recognition, IFN production and signaling, and ISGs [76]. Studies have shown that each viral protein can block different key signaling cascades. Viral RNA can be assembled with guanosine and methylated at the 5’ end by SARS-CoV-2 non-structural proteins, allowing the virus to efficiently escape recognition of the viral dsRNA by the host cell sensor [77]. Viral proteins inactivate key intermediaries in the IFN signaling cascade. A recent study showed that the SARS-CoV-2 ORF9b interacts with MAVS in mitochondria, resulting in a decrease in TRAF3 and TRAF6 [78]. The nsp13 and nsp15 proteins of SARS-CoV-2 interfere with TBK-1 signaling and activate IRF3 [78]. Another key virulence factor for SARS-CoV-2 is Nsp1, which inhibits the expression of the host gene; thus, it can effectively block the innate immune responses which could assist in the eradication of infection [71]. The right panel shows that the viral protein Nsp3 blocks IFN signaling by reversible post-translational modification of ISG 15 [79]. In patients with severe COVID-19, a significant mitigation of IFN I response is associated with a clinically persistent viral load and increased oxidative stress and an inflammatory response [70]. IFN: interferon; ISGs: interferon-stimulated genes; MAVS: mitochondrial antiviral signaling protein; nsp: nonstructural protein 13; TRAF: tumor necrosis factor receptor associated factor. Figure generated with Biorender (https://biorender.com/accessed on 6 September 2021).