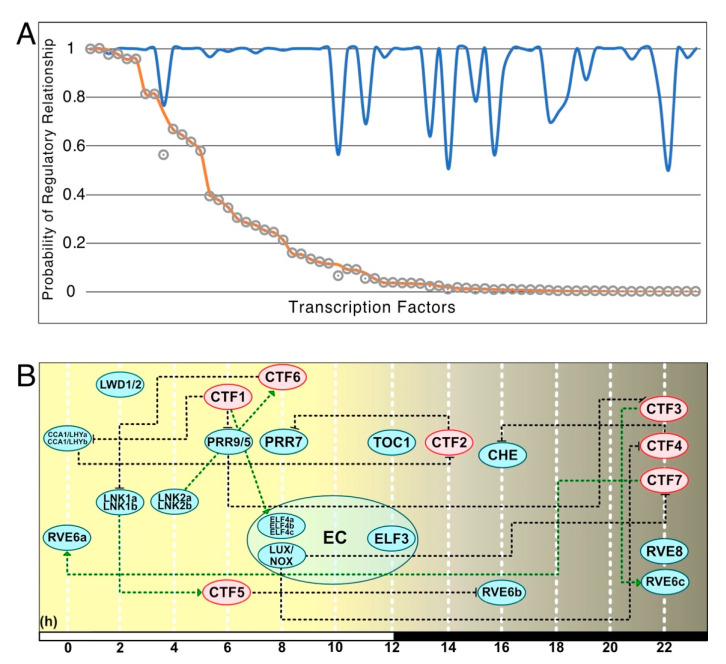
Figure 1.
Seven candidate core clock transcription factors (CTF1-7) are predicted to play a role in the core clock network of Kalanchoë fedtschenkoi. (A) Predicted regulatory relationships between candidate core clock transcription factors and known core clock genes via the Local Edge Machine [22]. Blue line is the probability that any known core clock transcription factor regulates a candidate clock transcription factor. Orange line is the probability that a candidate core clock transcription factor regulates any known core clock transcription factor. Grey circle is the probability of a regulatory relationship. (B) The seven candidate core clock transcription factors and their relationship with the core clock network in K. fedtschenkoi. Blue ovals represent core clock genes and red ovals represent candidate clock transcription factors. The numbers arrayed at the bottom of the image indicate the number of hours passed after first light. Green dotted lines represent activation of transcription and black dotted lines represent repression of transcription. White and black bars indicate daytime (12-hour) and nighttime (12-hour), respectively. CTF: Candidate core clock transcription factor, EC: Evening complex.