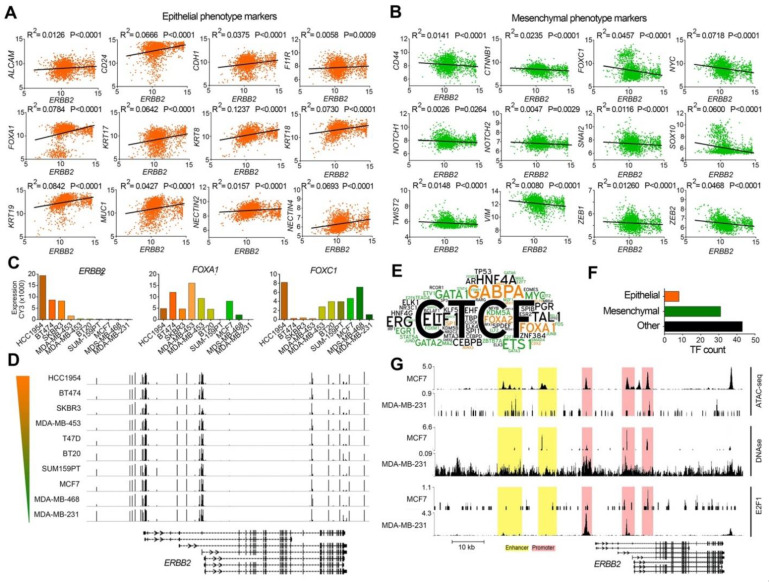
Figure 1.
Different ERBB2 expression and chromatin signature in epithelial-like and mesenchymal-like breast cancer cells. (A,B) Correlation between the mRNA expression of ERBB2 and epithelial (A) and mesenchymal (B) phenotype marker genes in human breast cancer tumors. The normalized RNA-seq expression data from 1904 breast tumors studied by METABRIC study [24] are presented by Z-score fold changes RNA-seq expression (v2 RSEM). (C) mRNA expression levels of ERBB2, FOXA1, and FOXC1 genes in 10 breast cancer cell lines. (D) Methylation levels of promoter CpG islands in the cell lines. The color gradient bar indicates HER2 expression level. Genomic coordinate: chr17:37,834,978–37,897,500 (GRCh37/hg19 assembly). (E) Word cloud diagram of transcription factors (TFs) detected bound to the ERBB2 gene at 10 kb upstream and downstream of motif Y. The different number of binding sites for each TF at the query region is illustrated as a different word size. TFs promoting epithelial and mesenchymal phenotypes are illustrated in orange and green colors respectively. (F) The number of the identified TFs based on their function in EMT. (G) ChIP-seq enrichment value peaks of ATAC-seq, DNase I hypersensitivity and E2F1 at ERBB2 gene regulatory regions in MCF7 and MDA-MB-231 cell lines. Genomic coordinate: chr17:39,643,771–39,735,523 (GRCh38/hg38 assembly).