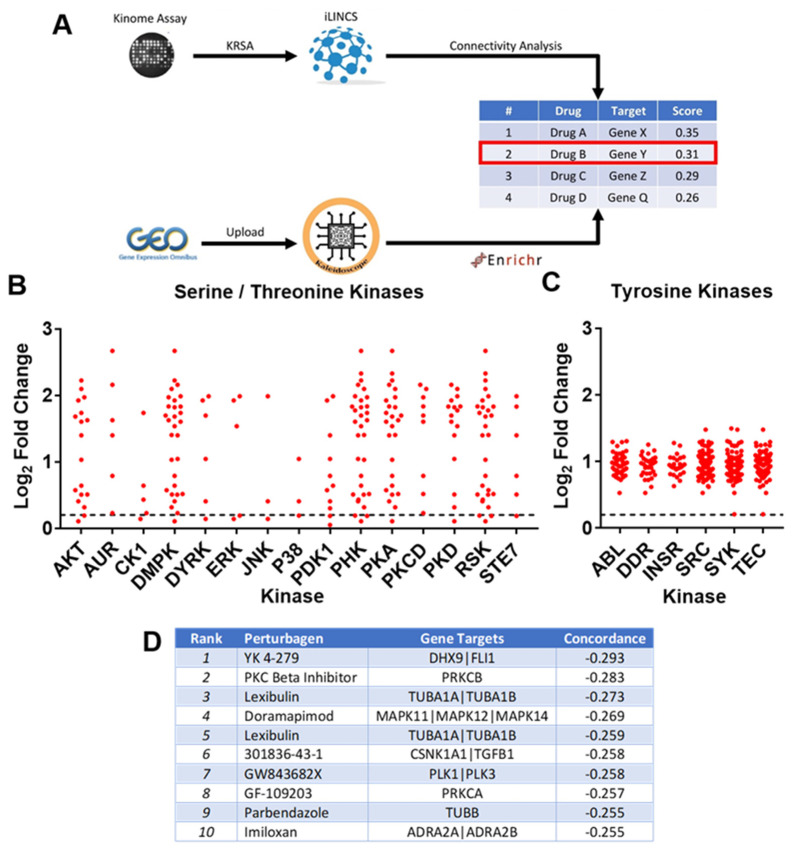
Figure 4.
Kinome profiling and in silico workflow for the identification of MC-LR-induced kinase activity and potential inhibitory compounds. (A) Schematic summarizing the overall workflow. Gene expression profiles derived from kinome profiles and published MC-LR exposure gene expression profiles were compared against perturbagen signatures in iLINCS to generate a list of hypothetical inhibitory compounds for the MC-LR-induced kinase activity. (B) Kinase activity from the serine/threonine kinase (STK) (C) and tyrosine kinase (PTK) arrays. (D) Identified hypothetical inhibitory compounds ranked by their inverse concordance with the MC-LR-induced signatures.