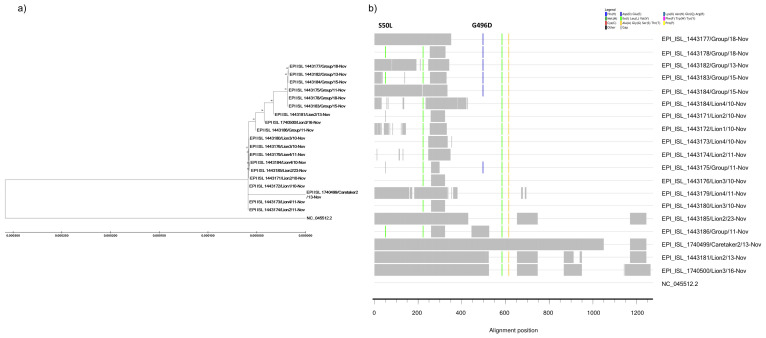
Figure 3.
Whole-SARS-CoV-2 genome sequencing and phylogenetic and comparative analyses. (a) Maximum likelihood phylogenetic analysis of the genomic sequences obtained from lion respiratory and fecal specimens at different time-points after infection was detected, as well as a nasopharyngeal sample from a lion caretaker, using the Hasegawa–Kishino–Yano model. The reference SARS-CoV-2/Wuhan-Hu-1 isolate (NCBI accession number: NC_045512.2) was included as an outgroup of the analysis. The tree with the highest log likelihood (−34294.32) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The phylogenetic tree is drawn to scale, with branch lengths measured in the number of substitutions per site. (b) Amino acid comparative sequence analysis of the spike protein of SARS-CoV-2. The panel shows mismatches (gray boxes) of the sequences obtained from lion respiratory and fecal specimens obtained throughout the study, as well as its respective caretaker, compared to the reference NC_045512.2 sequence. Comparative analyses determined the emerging mutations S50L and G496D specifically in fecal samples, whose positions have been pointed out in the plot. Colored lines in each position of the alignment show amino acid differences and sequencing gaps found among the different analyzed samples according to the legend. Abbreviations: C2, lion caretaker2; L1, Lion1; L2, Lion2; L3, Lion3; and L4, Lion4.