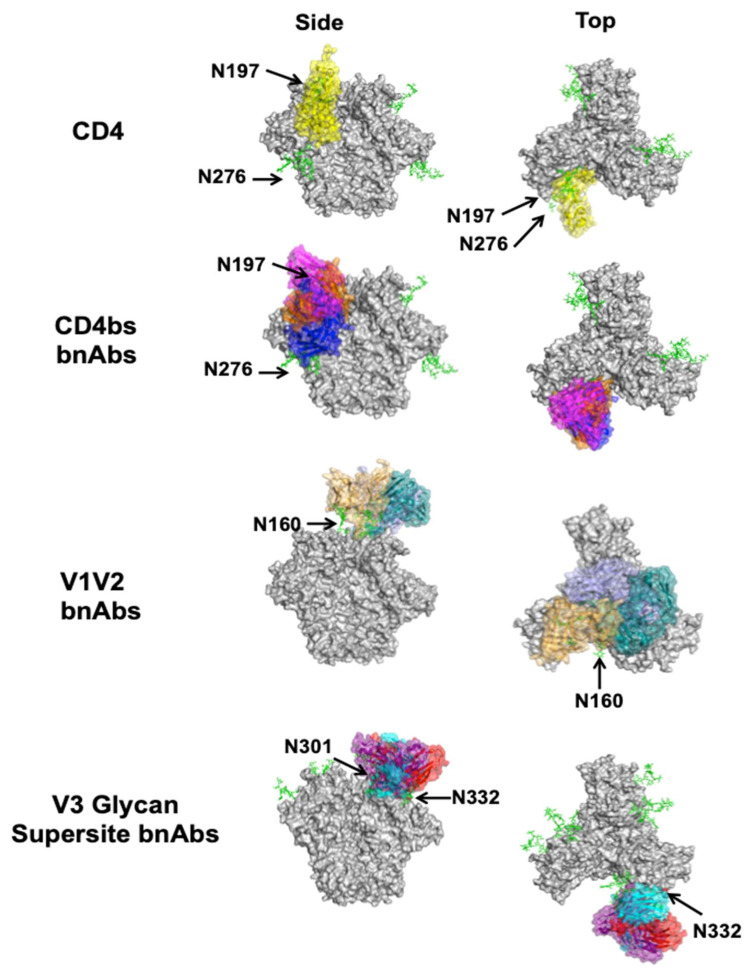
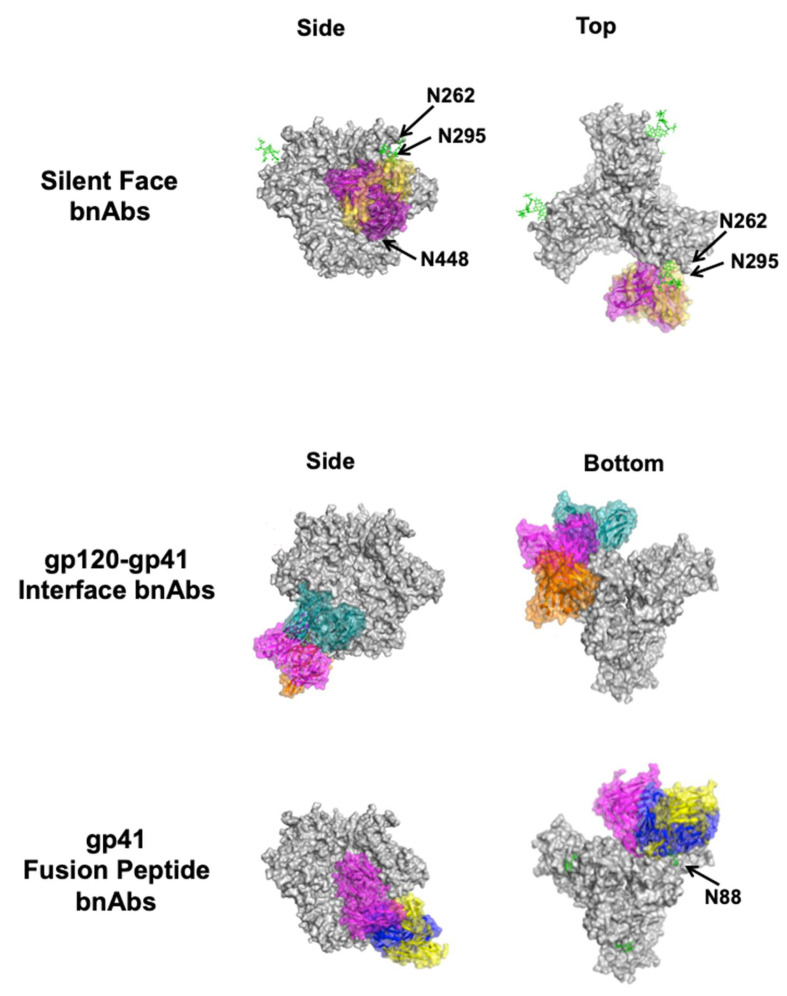
Figure 3.
Complexes between bnAbs and HIV-1 Env. Top and side views of CD4 (yellow, PDB ID: 1GC1) in complex with the Env trimer (gray, PDB ID: 5T3Z) are shown for reference along with CD4bs bnAbs VRC01 (blue, PDB ID: 4LST), CH103 (orange, PDB ID: 4JAN), and M1214-N1 (magenta, PDB ID: 6VY2) below. Of the V1V2 apex bnAbs structurally characterized to date, CAP256-VRC26.25 (teal, PDB ID: 6VTT), VRC38 (purple, PDB ID: 5VGJ), and PGT145 (orange, PDB ID: 5V8L) are shown. Of the V3 glycan supersite bnAbs structurally characterized to date, DH270.6 (cyan, PDB ID: 6UM6), 438-B11 (red, PDB ID: 6UTK), and Ab1485 (purple, 7KDE) are shown. The two known silent face bnAbs SF12 (magenta, PDB ID: 6OKP) and VRC-PG05 (yellow, PDB ID: 6BF4) are shown. Of the gp120–gp41 interface bnAbs structurally characterized to date, 8ANC195 (teal, PDB ID: 5CJX), 35O22 (magenta, PDB ID: 4TVP), and 1C2 (orange, PDB ID: 6P65) are shown. Only gp41 FP-directed bnAbs VRC34 (blue, PDB ID: 6NC3), ASC202 (yellow, PDB ID: 6NC2), and PGT151 (magenta, PDB ID: 6DCQ) are shown; MPER-directed bnAbs clash with a closed trimer and are therefore not shown. Relevant glycans are shown as green sticks and labeled according to the amino acid that is glycosylated. For clarity, only the Fv region of the antibodies is shown on a single protomer of the trimer, even if additional copies can bind. Side (left) and bottom or top (right) views are shown.