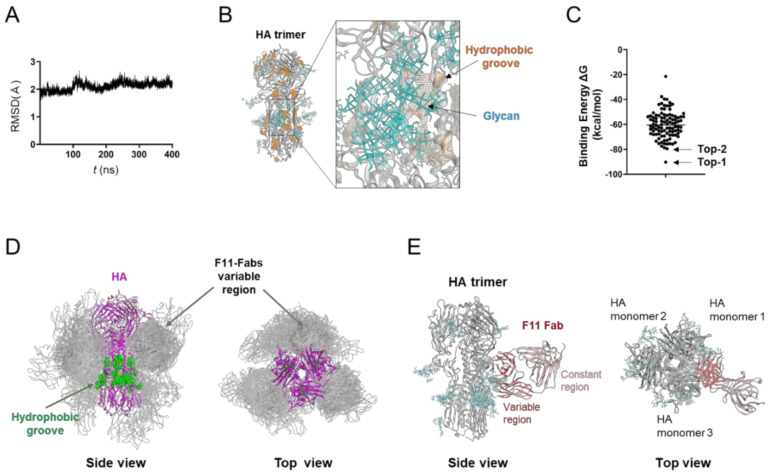
Figure 1.
Molecular modeling of the complex of glycosylated HA trimer and F11 Fab fragment. A three-dimensional model of the unliganded, glycosylated HA trimer ectodomain of A/Narita/1/2009 (H1N1)pdm09 virus [19] was constructed by the homology modeling method using the Molecular Operating Environment (MOE) (Chemical Computing Group Inc., Montreal, QC, Canada), glycosylated using tools in GLYCAM-Web [21], and subjected to MD simulation using modules in the Amber 16 program package [23]. The HA trimer model in the equilibrium state under solution conditions was used for the docking simulations of the F11 Fab fragment using the Dock application of MOE [29,30]. (A) Root mean square deviation (RMSD) [23] between the structure of the initial HA structure and those at given time points of the MD simulation of glycosylated HA trimers calculated using the cpptraj module in AmberTools 16 as described previously [22]. (B) Side view of the HA trimer model at 400 ns of MD simulation. (C) Distribution of binding energies among the top 100 binding poses generated using the Dock application of MOE [29,30]. (D) Superposition of 100 docking poses. The purple region indicates the HA trimer. Green regions indicate the hydrophobic groove [50] in the HA trimer [53,54]. Grey regions indicate F11 Fab variable regions of the top 100 docking poses. Glycans attached on the HA trimer are not shown here in order to highlight the binding mode of the F11 Fab variable region. (E) The second highest ranked binding mode of F11 Fab (top-2 binding mode). Cyan branches indicate the high-mannose oligosaccharide Man5GlcNAc2.