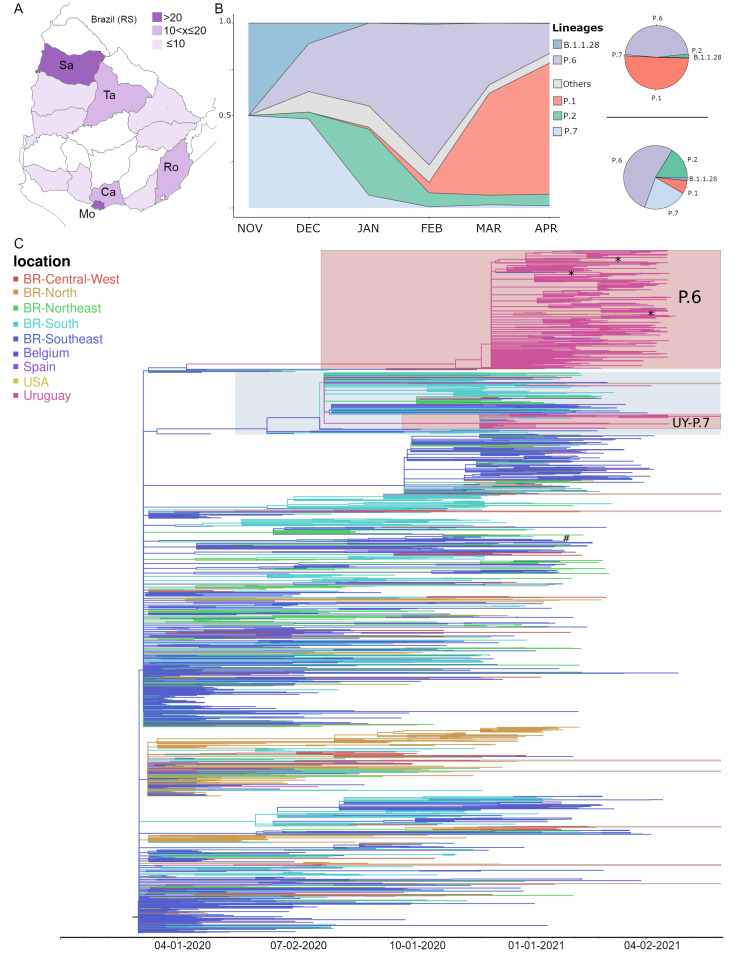
Figure 1.
Geographic distribution, prevalence, and maximum likelihood analysis of P.6. (A) Map of Uruguay showing the number of sequences classified as P.6 (B.1.1.28 + Q675H + Q677H) in every department (n = 165 for samples with known geographical source in Uruguay). Department labels are as follows: Sa (Salto), Ta (Tacuarembo), Ro (Rocha), Ca (Canelones), Mo (Montevideo). The metropolitan area corresponds to Mo and Ca. The border with Brazil is shown. RS stands for Rio Grande do Sul, the southernmost Brazilian state. (B) To the left, Pango lineage proportions of all available Uruguayan samples calculated monthly, from November 2020 to April 2021. The “Others” category includes B.1, B.1.1, B.1.1.1, B.1.1.34, B.1.177, B.1.177.12, and B.1.238. To the right, pie charts show Pango lineage prevalence in the metropolitan area (top) versus the rest of the country (bottom). (C) Maximum likelihood phylogeographic analysis of lineage B.1.1.28 samples (n = 1787) from Uruguay (n = 355); Brazil (n = 1428); and the USA, Spain, and Belgium (n = 4) inferred by an ancestral character reconstruction method implemented in PastML. Tips and branches are colored according to sampling location and the most probable location state of their descendent nodes, respectively, as indicated in the legend. Shaded boxes highlight the major B.1.1.28 clades in Uruguay. P.6 is the assigned Pango name for the clade B.1.1.28 + Q675H + Q677H discussed here, while UY-P.7 (corresponding to UYP7 in the main text) is a B.1.1.28 clade carrying mutation N:P13L widely distributed in southern Brazil (recently assigned as P.7). Brazilian P.7 is shown with a gray shadow. Asterisks (*) indicate the sequences dispersed from Uruguay to the USA and Spain. The hash indicates the B.1.1.28 + Q675H + Q677H sample from Belgium. The time-scaled tree was rooted with the earliest sequence (collection date 5 March 2020). Branch lengths are drawn to scale indicating nucleotide substitutions per site per year.