Figure 1. p53 regulates glucose, lipid, amino acid, nucleotide, and iron metabolism.
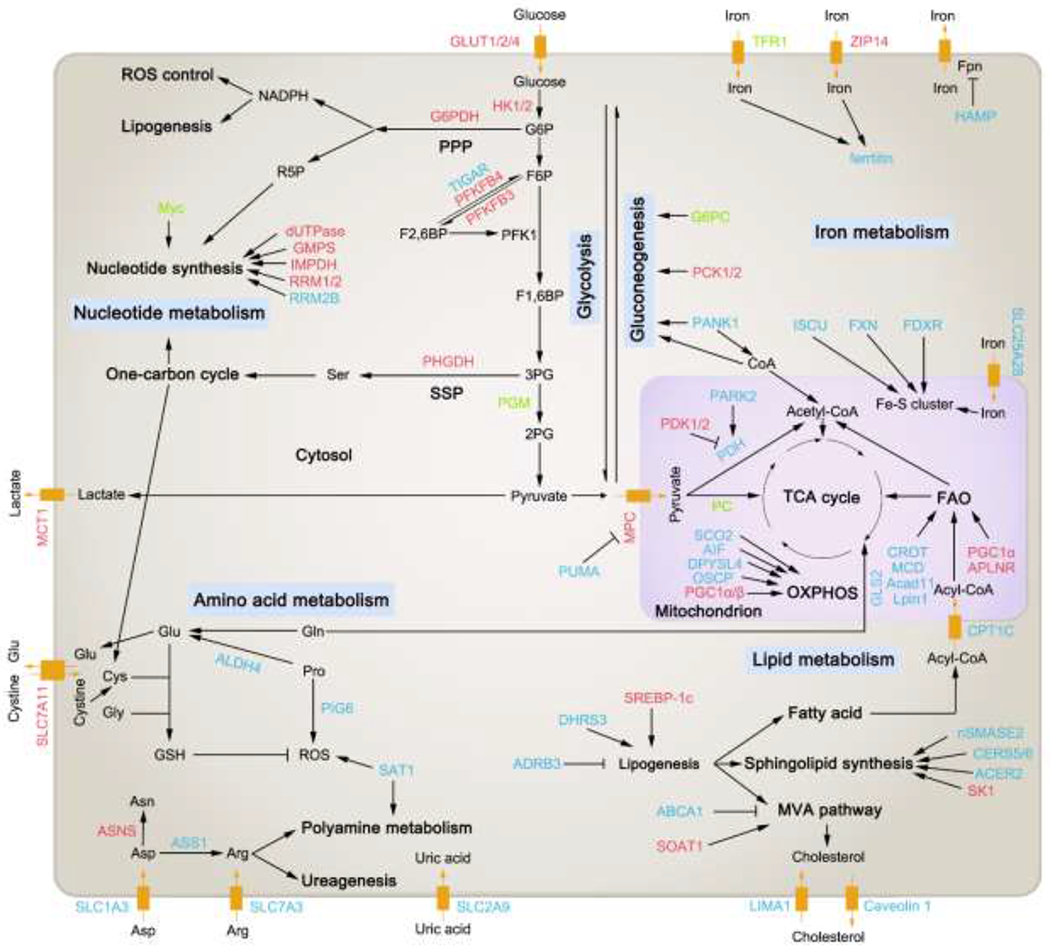
p53 participates in the regulation of metabolism of diverse biomolecules (including glucose, lipid, amino acid, nucleotide, and iron) in a transcription factor (TF)-dependent or –independent way. Major target genes regulated by p53 in these metabolic pathways are shown in this figure. For the full names of them, please refer to Table 1. Those proteins that are modulated positively by p53 (directly or indirectly), like promoting their expressions or activities, are depicted in blue. Oppositely, those proteins that are modulated negatively by p53 (directly or indirectly), like suppressing their expressions or activities, are depicted in red. If some protein can either be promoted or suppressed by p53 in different circumstances, it is depicted in green. Black arrows indicate positive effects. Black perpendicular bars indicate negative effects. G6P, glucose-6-phosphate; PPP, pentose phosphate pathway; F6P, fructose-6-phosphate; F2,6BP, fructose-2,6-bisphosphate; PFK1, phosphofructokinase 1; F1,6BP, fructose-1,6-bisphosphate; 3PG, 3-phosphoglycerate; SSP, serine synthesis pathway; 2PG, 2-phosphoglycerate; Fpn, ferroportin; Ser, serine; Glu, glutamate; Gln, glutamine; Cys, cysteine; Gly, glycine; Pro, proline; Asp, asparate; Asn, asparagine; Arg, arginine; GSH, glutathione; TCA cycle, tricarboxylic acid cycle; OXPHOS, oxidative phosphorylation; MVA pathway, mevalonate pathway.
