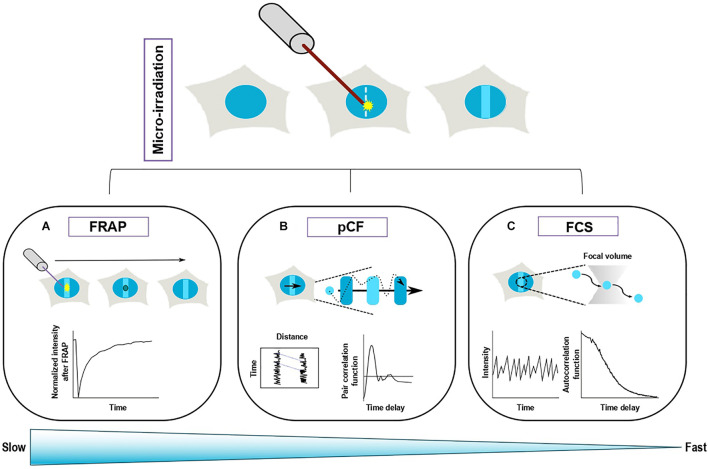
FIGURE 2.
Complementary fluorescence-based methods to study the turnover of repair proteins at sites of DNA damage. Top panel: Recruitment of a fluorescently tagged protein at sites of DNA damage induced by laser irradiation. Bottom panel: Three different methods allow to monitor protein turnover at sites of damage. (A) Fluorescence recovery after photobleaching (FRAP) is based on the photobleaching of a sub-region (solid circle) of the area of DNA damage. The fluorescence recovery within the bleached area gives access to protein turnover at DNA lesions. (B) Pair correlation function (pCF) assesses the movements of the proteins within a line-scan across the damage region. The fluorescence signals from two different pixels along the acquisition line are cross-correlated, which allows to estimate the characteristic transit time between these two pixels. (C) Fluorescence Correlation spectroscopy (FCS) collects the fluctuations arising from the movement of fluorescently tagged proteins in and out of a confocal volume located within the DNA damage region. The characteristics of the protein turnover at sites of damage are derived from the analysis of the autocorrelation of the fluorescence fluctuations. FRAP is more appropriate to assess slow turonver while FCS allows to study fast protein exchange.