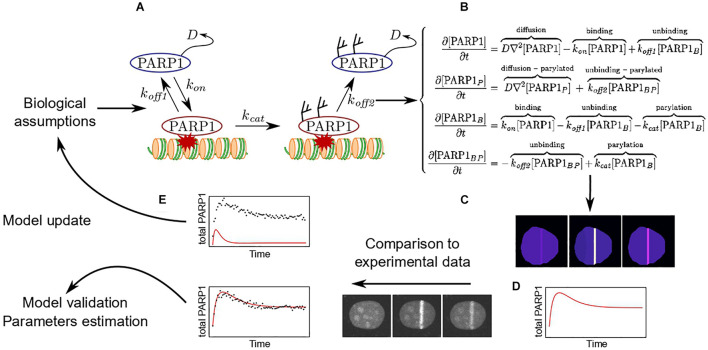
FIGURE 3.
Development of a mathematical model to assess PARP1 dynamics at DNA lesions. (A) Based on specific biological assumptions, it is possible to build a model describing the different states of PARP1 in and out the DNA damage area as well as the exchange rates between these different states. (B) The model can be transcribed in a set of reaction-diffusion differential equations. (C) Solving the set of equations allows to predict the evolution of the spatio-temporal distribution of PARP1 within the nucleus in response to induction of DNA damage. (D) Based on this simulation, it is possible to obtain predicted recruitment kinetic curves. (E) These simulated recruitment curves can be fitted to the experimental ones. If the model is unable to fit the experimental data, a new model needs to be rebuilt. In contrast, if the model allows to properly fit the data, it is possible to estimate the parameters included in the model.