Figure 1. ROS‐associated functional pathways changes in aging human and Fischer 344 rat hearts.
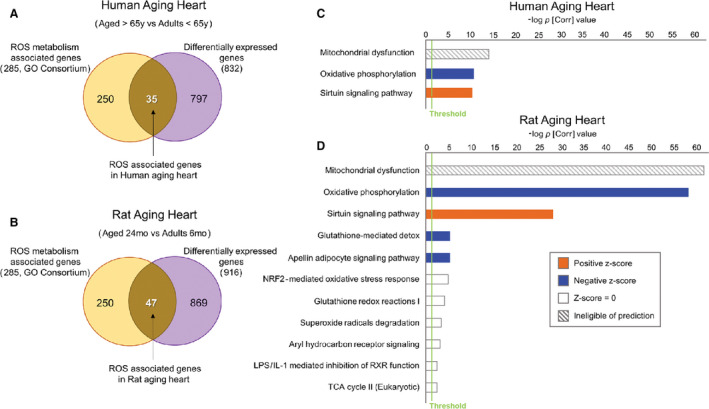
A, Out of the 832 differentially expressed genes (DEG) in aged compared to adult hearts, 35 genes (4.2%) were associated with ROS metabolism, with 27 genes downregulated and 8 upregulated in the human aged heart. B, In the aging rat heart, out of 916 DEGs we found 47 (5.1%) ROS‐metabolism‐associated genes (39 downregulated, 8 upregulated). C, Functional and pathway analysis exposed three significant canonical pathways enriched in the human dataset: “mitochondrial dysfunction” (white/gray pattern=ineligible of pathway prediction), “oxidative phosphorylation” (blue=predicted decrease in activity of pathway), and “sirtuin signaling pathways” (orange=predicted increase in activity). Threshold was set at P [Corr] <0.05 (green line). D, ROS‐pathway‐associated genes from aged rats enriched distinctive and significant functional categories that were similar in rats and humans for the top three canonical pathways, with stronger significance reported for the former. Furthermore, “glutathione‐mediated detox” and “apelin adipocyte signaling pathway” also predicted decrease in activity (blue bars). Six other functional pathways were significantly enriched; however, they showed neutral predicted activity. (For details, see Methods and Materials). IPA indicates Ingenuity Pathways Analysis; P [Corr], Corrected P value including multiple testing correction (Benjamini Hochberg false discovery rate); and ROS, reactive oxygen species.
