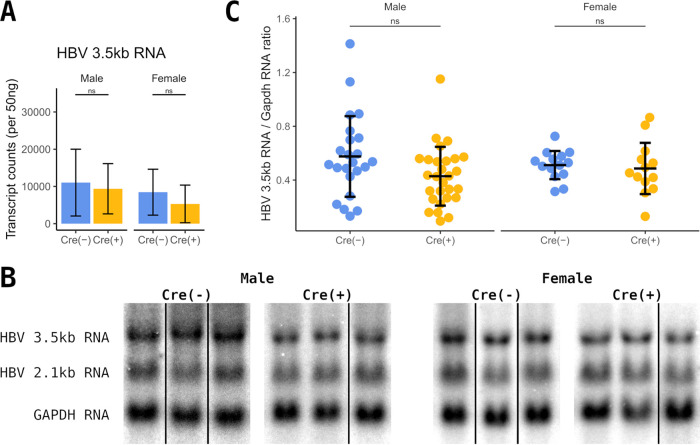
FIG 6.
Analysis of HBV RNA expression in liver-specific β-catenin-null HBV transgenic mice. (A) Counts of HBV 3.5-kb transcripts in total liver RNA from control (Cre−) and constitutive liver-specific β-catenin-null HBV transgenic mice (Cre+) by NanoString gene expression analysis (n = 9 per group). (B) RNA filter hybridization analysis on total liver RNA from control (Cre−) and constitutive liver-specific β-catenin-null HBV transgenic mice (Cre+). The glyceraldehyde 3-phosphate dehydrogenase (Gapdh) transcript was used as an internal control for the quantitation of the HBV 3.5-kb RNA. Noncontiguous lanes from a single analysis are presented. (C) Quantitation of viral 3.5-kb RNA from the RNA filter hybridization analysis. Means and standard deviations are presented (male Cre−, n = 23; male Cre+, n = 28; female Cre−, n = 14; female Cre+, n = 13). Statistically significant differences between HBV transgenic mice by Wilcoxon rank sum test are indicated; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.