ABSTRACT
Cleavage of the influenza A virus (IAV) hemagglutinin (HA) by host proteases is indispensable for virus replication. Most IAVs possess a monobasic HA cleavage site cleaved by trypsin-like proteases. Previously, the transmembrane protease TMPRSS2 was shown to be essential for proteolytic activation of IAV HA subtypes H1, H2, H7, and H10 in mice. In contrast, additional proteases are involved in activation of certain H3 IAVs, indicating that HAs with monobasic cleavage sites can differ in their sensitivity to host proteases. Here, we investigated the role of TMPRSS2 in proteolytic activation of avian HA subtypes H1 to H11 and H14 to H16 in human and mouse airway cell cultures. Using reassortant viruses carrying representative HAs, we analyzed HA cleavage and multicycle replication in (i) lung cells of TMPRSS2-deficient mice and (ii) Calu-3 cells and primary human bronchial cells subjected to morpholino oligomer-mediated knockdown of TMPRSS2 activity. TMPRSS2 was found to be crucial for activation of H1 to H11, H14, and H15 in airway cells of human and mouse. Only H9 with an R-S-S-R cleavage site and H16 were proteolytically activated in the absence of TMPRSS2 activity, albeit with reduced efficiency. Moreover, a TMPRSS2-orthologous protease from duck supported activation of H1 to H11, H15, and H16 in MDCK cells. Together, our data demonstrate that in human and murine respiratory cells, TMPRSS2 is the major activating protease of almost all IAV HA subtypes with monobasic cleavage sites. Furthermore, our results suggest that TMPRSS2 supports activation of IAV with a monobasic cleavage site in ducks.
IMPORTANCE Human infections with avian influenza A viruses upon exposure to infected birds are frequently reported and have received attention as a potential pandemic threat. Cleavage of the envelope glycoprotein hemagglutinin (HA) by host proteases is a prerequisite for membrane fusion and essential for virus infectivity. In this study, we identify the transmembrane protease TMPRSS2 as the major activating protease of avian influenza virus HAs of subtypes H1 to H11, H14 and H15 in human and murine airway cells. Our data demonstrate that inhibition of TMPRSS2 activity may provide a useful approach for the treatment of human infections with avian influenza viruses that should be considered for pandemic preparedness as well. Additionally, we show that a TMPRSS2-orthologous protease from duck can activate avian influenza virus HAs with a monobasic cleavage site and, thus, represents a potential virus-activating protease in waterfowl, the primary reservoir for influenza A viruses.
KEYWORDS: influenza virus, TMPRSS2, hemagglutinin, monobasic cleavage site, virus-activating protease, primary airway cells, avian influenza, morpholino oligomers
INTRODUCTION
Influenza A virus (IAV) causes highly contagious infection in various avian and mammalian species, including humans, pigs, and poultry, and is recognized as an important zoonotic infectious agent with pandemic potential. IAV belongs to the family Orthomyxoviridae and is an enveloped virus with a negative-sense, single-stranded RNA genome that consists of 8 segments encoding up to 17 proteins. Based on antigenic characteristics of the two envelope glycoproteins, hemagglutinin (HA) and neuraminidase (NA), avian IAVs are divided into 16 distinct HA and 9 NA subtypes. Two additional HA and NA subtypes, H17N10 and H18N11, have been described in bats (reviewed in reference 1). Wild birds, primarily ducks, gulls, and shorebirds, are the natural host and primary reservoir for IAVs and may transmit IAV to other host species (2).
In humans, IAVs cocirculate with influenza B viruses (IBVs) with varying predominance and are responsible for seasonal outbreaks of acute respiratory disease (flu), with 3 to 5 million cases of severe respiratory illness and 290,000 to 650,000 deaths worldwide (WHO, November 2018). In wild birds, IAV infections are usually asymptomatic, with virus replication taking place primarily in epithelial cells of the intestinal tract and high virus load in feces (reviewed in references 3 and 4). Transmission of avian IAV to domestic poultry may lead to transformation into highly pathogenic IAV (HPAIV), resulting in large outbreaks of disease in poultry in many countries worldwide, accompanied by significant economic losses to the poultry industry and posing a potential threat to both animal and human public health. Infections of low pathogenic avian IAV (LPAIV) in domestic poultry cause primarily mild respiratory disease and lowered egg production, whereas HPAIVs cause severe systemic infection with up to 100% mortality (fowl plague or bird flu). Occasionally avian IAVs that circulate in poultry cross the species barrier and infect humans, with variable consequences (5–8). Importantly, emergence of novel IAV with different HA in a naive human population poses the risk of initiating a pandemic, as exemplified by the four influenza pandemics in recent history. The most devastating influenza pandemic, the 1918 Spanish flu, resulted in an estimated 20 to 50 million deaths worldwide. Furthermore, within the last 2 decades, human infections with avian IAV caused by exposure to infected poultry have attracted more attention and are frequently reported. In 1997, an HPAIV of subtype H5N1 emerged in Hong Kong and caused an outbreak with 18 human cases of severe respiratory disease, 6 of whom died (reviewed in reference 9). The H5N1 virus reemerged in 2003 in humans, and a total number of 862 human infections and 455 deaths have been reported to date (WHO, December 2020). In 2013, an H7N9 virus emerged in China and has caused a total of 1,568 human cases, including 616 deaths (WHO, May 2021). Serological and virological surveillance studies reveal that infections with H6, H7, H9, and H10 LPAIVs as well as H5N6, H5N8, and H7 HPAIVs also are occurring with increased frequency and are associated with severe or even fatal disease (5, 7, 10).
IAV replication is initiated by the major surface glycoprotein HA through its binding to sialic acid-containing receptors and mediating fusion of the viral lipid envelope with the endosomal membrane. HA is synthesized as a fusion-incompetent precursor, HA0, that must be cleaved by a host cell protease into the subunits HA1 and HA2 to prime HA for membrane fusion (reviewed in reference 11). Cleavage of HA has long been known to be a major determinant of avian IAV pathogenicity in poultry. HPAIVs of subtypes H5 and H7 possess a multibasic cleavage site of the consensus sequence R-X-R/K-R↓ (arrow indicates cleavage) that is processed by ubiquitous furin and proprotein convertase 5/6 (PC5/6), supporting systemic spread of infection (reviewed in reference 12). In contrast, the HAs of LPAIVs of the 16 HA subtypes known to date in waterfowl possess a single arginine residue, designated monobasic cleavage site, which is not cleaved by furin but must be activated by trypsin-like serine proteases instead. To date, the proteases that carry out HA cleavage at a monobasic cleavage site in birds have not been clearly identified but are believed to be restricted to the respiratory and intestinal tract, confining viral tropism to those tissues (reviewed in reference 12). HPAIV may emerge from LPAIV by acquisition of a multibasic cleavage site (reviewed in reference 13).
Mammalian IAVs, including human viruses of subtypes H1, H2, and H3, and IBVs possess an HA with a monobasic cleavage site. In 2006, we identified the type II transmembrane serine protease TMPRSS2 as a human airway protease that is able to cleave a monobasic HA cleavage site in vitro (14). More recent studies by us and others demonstrated that TMPRSS2 is essential for proteolytic activation and spread of human IAVs of subtype H1 and H2, zoonotic H7N9 virus, and avian H10 virus in mice (15–19). Importantly, mice deficient in TMPRSS2 expression were resistant to pathogenesis resulting from virus infection. In contrast, activation of some human H3N2 viruses and IBV was found to be independent of TMPRSS2 in mice and supported by alternative proteases (15–17, 20, 21). Together, these previous studies have made clear that activation of HA is essential for virus spread and pathogenesis in mammalian hosts. Furthermore, these studies have established that various HAs having a monobasic cleavage site can differ in their sensitivity to host cell proteases in mice in vivo. More recently, we elucidated the role of TMPRSS2 in IAV activation in human airway cells (22). We demonstrated that TMPRSS2 is the major HA-activating protease of both human H1 and H3 and zoonotic H7N9 IAVs in the human airway cell line Calu-3, in primary human bronchial epithelial cells (HBEC), and primary human type II alveolar epithelial cells (AECII).
In the present study, we investigated whether TMPRSS2 does support proteolytic activation of avian IAV HAs of different subtypes in airway cells of mammalian origin. To this end, we generated a panel of IAV reassortants containing representative avian HAs of subtype H1 to H11 and H14 to H16 in the background of the avian- and mammalian-adapted strain SC35M (23). We then studied HA activation and multicycle replication of those reassortant viruses in lung explants from TMPRSS2-deficient mice as well as in Calu-3 cells and primary HBEC cultures with peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO)-induced knockdown of TMPRSS2 activity. Beyond that, we cloned an avian TMPRSS2-orthologous protease from duck lung tissue, transiently expressed it in MDCK cells, and examined its proteolytic activation of HAs of almost all HA subtypes.
RESULTS
Reassortant SC35M IAVs bearing HAs of different subtypes.
To study the TMPRSS2-mediated proteolytic activation of IAVs with different HA subtypes in murine and human airway cells, we generated a panel of 1:7 reassortant viruses that carry representative avian HAs of different subtypes described previously (24) and shared the other seven gene segments from the avian- and mouse-adapted strain SC35M (23). SC35M was obtained by serial passages of A/seal/Massachusetts/1/1980 (H7N7) in chicken embryo fibroblasts and subsequently in mice (23). Since no H5 of an LPAIV was available for this study, we used the reassortant H5N1-PR8 that contains the HA and NA of an avian H5N1 virus isolated from a human (A/Vietnam/1203/2004) in the genetic background of PR8 (25). Its original multibasic HA cleavage site (PQRERRRKKR↓) was converted by mutagenesis to a monobasic one. For H7, we used the H7-PR8 reassortant possessing the HA of A/Anhui/1/13 (H7N9) with a monobasic cleavage site in the PR8 background, described previously (22). The donors of the HAs and abbreviations of the reassortant viruses are summarized in Table 1. An amino acid sequence alignment of all HAs is provided as Fig. S1 in the supplemental material.
TABLE 1.
List of wild-type viruses and recombinant viruses including HA cleavage site sequences used in this study
| Virus | Abbreviation | Cleavage site motifa |
|---|---|---|
| HA donor for recombinant virus | ||
| A/duck/Bavaria/1/1977 (H1N1) | H1-SC35M | PSIQSR↓G |
| A/sentinel mallard/Germany/S/Ra517K/2007 (H2N5) | H2-SC35M | PQIESR↓G |
| A/duck/Ukraine/1/1963 (H3N8) | H3-SC35M | PEKQTR↓G |
| A/mallard/Germany/1240/1/2007 (H4N6) | H4-SC35M | PEKASR↓G |
| A/Vietnam/1203/2004 (H5N1) | H5N1-PR8 | PQIETR↓G |
| A/turkey/Germany/R617/2007 (H6N2) | H6-SC35M | PQIENR↓G |
| A/Anhui/1/2013 (H7N9) | H7-PR8 | PEIPKGR↓G |
| A/turkey/Ontario/6118/1968 (H8N4) | H8-SC35M | PSVEPR↓G |
| A/chicken/Emirates/R66/2002 (H9N2) | H9-SC35M | PARSSR↓G |
| A/mallard/Germany/R2075/2007 (H10N7) | H10-SC35M | PEIMQGR↓G |
| A/domestic duck/Germany/R784/2006 (H11N1) | H11-SC35M | PAIASR↓G |
| A/mallard/Gurijev/263/1982 (H14N3) | H14-SC35M | PGKQA K ↓G |
| A/shearwater/West Australia/2576/1979 (H15N9) | H15-SC35M | PEKIRTR↓G |
| A/black-headed gull/Sweden/5/1999 (H16N3) | H16-SC35M | PSVGER↓G |
| Wild-type viruses | ||
| A/Victoria/3/1975 (H3N2) | H3N2/Victoria | PEKQTR↓G |
| A/HongKong/1/1968 (H3N2) | H3N2/HongKong | PEKQTR↓G |
| A/quail/Shantou/782/2000 (H9N2) | H9N2/Shantou | PARSSR↓G |
| A/turkey/Wisconsin/1/1966 (H9N2) | H9N2/Wisconsin | PAVSSR↓G |
| B/Malaysia/2506/2004 | IBV | PAKLLKER↓G |
The arrow indicates the place of proteolytic cleavage. Basic amino acids crucial for HA cleavage by relevant proteases are shown in boldface.
Proteolytic activation of avian HAs in lung cells of TMPRSS2-deficient mice.
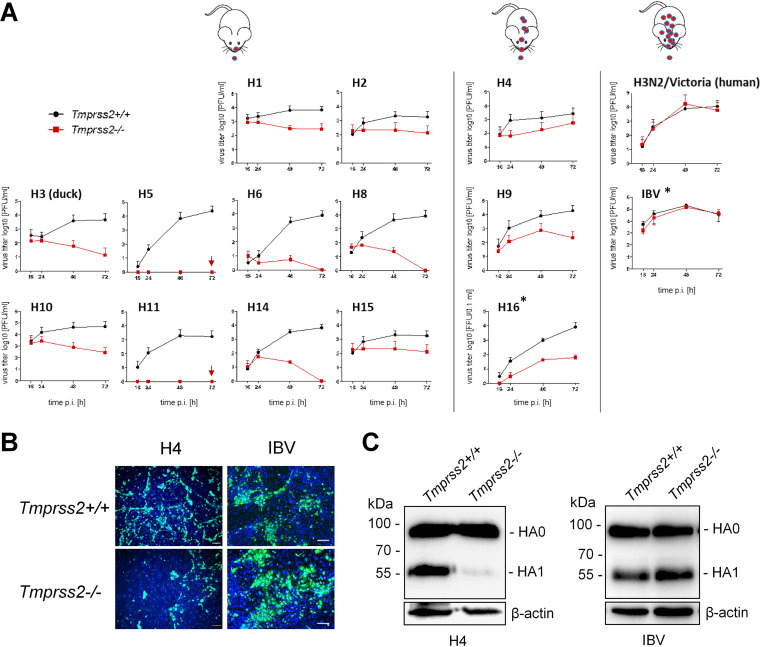
To investigate the role of TMPRSS2 in activation and multicycle replication of different avian HA subtypes in murine airway cells, we studied growth kinetics of the HA-SC35M reassortants containing avian HAs of subtypes H1, H2, H3, H4, H6, H8, H9, H10, H11, H14, and H15 and H5N1-PR8 in lung explants of TMPRSS2-deficient mice (Tmprss2−/−) and wild-type animals (Tmprss2+/+). IAV HAs of subtype H12 and H13 were not available for this study. Explants were inoculated with virus at a low multiplicity of infection (MOI) and incubated for 72 h. Virus titers in the supernatants were determined by plaque assay at different time points postinfection (p.i.). As shown in Fig. 1A, multicycle replication of reassortants of HA subtypes H1, H2, H5, H6, H8, H10, H11, H14, and H15 was strongly suppressed in lung explants of Tmprss2−/− mice compared to their efficient growth in lung tissue of wild-type animals. Interestingly, multicycle replication of H3-SC35M possessing HA of A/duck/Ukraine/1/1963 (H3N8) was strictly TMPRSS2 dependent in murine lung explants. In contrast, the human virus H3N2/Victoria replicated to similar titers in explants of both wild-type and TMPRSS2-deficient mice, similar to what has been observed for other human H3N2 viruses previously (15–17).
FIG 1.
Multicycle replication analysis of IAVs of different HA subtypes in lung cells of TMPRSS2-deficient mice. (A) Lung explants of TMPRSS2-deficient mice (Tmprss2−/−) or wild-type mice (Tmprss2+/+) were inoculated with 4 × 103 PFU of the indicated virus and incubated for 72 h. Virus titers were determined at 16, 24, 48, and 72 h postinfection (p.i.) by plaque assay. Replication of H5N1-PR8 and H11-SC35M was completely blocked in lung explants of TMPRSS2−/− mice (indicated by a red arrow). Panels showing growth kinetics of H16-SC35M and IBV in primary type II alveolar epithelial cells (AECII) of Tmprss2+/+ and Tmprss2−/− mice are indicated by asterisks. Murine AECII were inoculated with H16-SC35M or IBV at an MOI of 0.01 and incubated for 72 h. At the indicated time points, virus titers were determined by focus formation assay. Data are mean values ± standard deviations (SD) from two or three independent experiments. Schematic mice illustrate virus activation (red symbols) and spread along the airways in the absence of TMPRSS2. (B) Multicycle replication of H4-SC35M and IBV in AECII isolated from Tmprss2+/+ and Tmprss2−/− mice. Cells were cultivated and inoculated with H4-SC35M or IBV at an MOI of 0.01 and incubated for 24 h. Subsequently AECII were fixed, permeabilized, and immunostained for NP (green). The nuclei were stained using DAPI (blue). Representative images from one (H4-SC35M) or three (IBV) independent experiments are shown. Scale bars indicate 100 μm. (C) Analysis of HA cleavage in AECII of Tmprss2+/+ and Tmprss2−/− mice. Cells were inoculated with H4-SC35M or IBV at an MOI of 0.8 and 0.04, respectively, and incubated for 24 h. Cell lysates were subjected to SDS-PAGE and Western blotting with HA-specific antibodies. Beta-actin served as a loading control. HA2 was not detected by the HA-specific antibodies.
H9-SC35M, possessing the dibasic cleavage site motif R-S-S-R↓, was able to replicate in lung explants of Tmprss2−/− mice, although to much lower titers than virus replication in explants of Tmprss2+/+ mice (Fig. 1A). These data suggest that another protease can promote H9 HA activation in murine lung cells but cannot fully compensate for the lack of TMPRSS2 activity in H9 HA cleavage. A slight increase in virus titer compared to the inoculum was also observed for H4-SC35M in lung explants of Tmprss2−/− mice at 48 and 72 h p.i., although the effect was less pronounced than that in the case of H9-SC35M. We also examined proteolytic activation of H16 HA in lung cells of Tmprss2−/− and Tmprss2+/+ mice. H16 HA has been shown to contain an unusual ɑ-helix in the cleavage site that hides the arginine residue and is resistant to cleavage by trypsin in vitro (26). Murine lung explants were not available for this experiment. We therefore used primary type II alveolar epithelial cells (AECII) instead, as they have been demonstrated to provide a suitable model to study HA cleavage by different host proteases in murine lung (22, 27). Multicycle replication of H16-SC35M was reduced in AECII of TMPRSS2-deficient mice; however, virus growth was not fully blocked, indicating that H16 HA is cleaved to some extent by proteases other than TMPRSS2 in murine lung cells. IBV that was used as a control replicated efficiently in AECII of both TMPRSS2-deficient and wild-type mice, as described previously (22).
To confirm that inhibition of virus multiplication in lung cells of Tmprss2−/− mice is specifically caused by inhibition of activation of HA, we analyzed HA cleavage in lung cells of Tmprss2−/− and Tmprss2+/+ mice. Since only small amounts of HA were detectable in lung explants infected with the different viruses (data not shown), we used AECII for HA cleavage analysis. AECII of Tmprss2−/− and Tmprss2+/+ mice were inoculated either with H4-SC35M or IBV at a low MOI for analysis of virus spread or at a high MOI for HA cleavage analysis. At 24 h p.i., cells were fixed and immunostained using antibodies against the viral nucleoprotein (NP), and lysates were subjected to SDS-PAGE under reducing conditions and Western blot analysis with H4- or IBV-HA-specific antibodies. As shown in Fig. 1B, efficient spread of H4-SC35M was visible in AECII of Tmprss2+/+ mice, whereas spread in AECII of Tmprss2−/− mice was strongly reduced, similar to what has been observed in lung explants. Consistent with this, efficient cleavage of H4 HA0 into HA1 and HA2 (HA2 not detected by the antibody) was observed in primary AECII of Tmprss2+/+ mice, while cleavage of H4 HA0 was strongly inhibited in AECII of Tmprss2−/− mice (Fig. 1C). The data demonstrate that TMPRSS2 is crucial for efficient H4 cleavage in murine lung cells. Detection of small amounts of cleaved HA1, however, indicated that H4 is cleaved with low efficiency by an unknown protease(s). IBV was able to undergo multicycle replication in AECII of both Tmprss2−/− and Tmprss2+/+ mice, and HA0 was efficiently cleaved, as described previously (22).
Together, the data demonstrate that TMPRSS2 is essential for activation of avian H1, H2, H3, H5, H6, H8, H10, H11, H14, and H15 HA, having a monobasic cleavage site in murine lung cells. Further, the data show that H9 HA with R-S-S-R↓ cleavage site, H16 HA, and, to a lesser extent, H4 HA can be activated by an unknown protease(s) in addition to TMPRSS2 in murine lung, although with reduced efficiency. Thus, our findings suggest that TMPRSS2-independent activation is unique to certain human H3 viruses and IBV in mice.
Role of TMPRSS2 in proteolytic activation of HAs in Calu-3 cells.
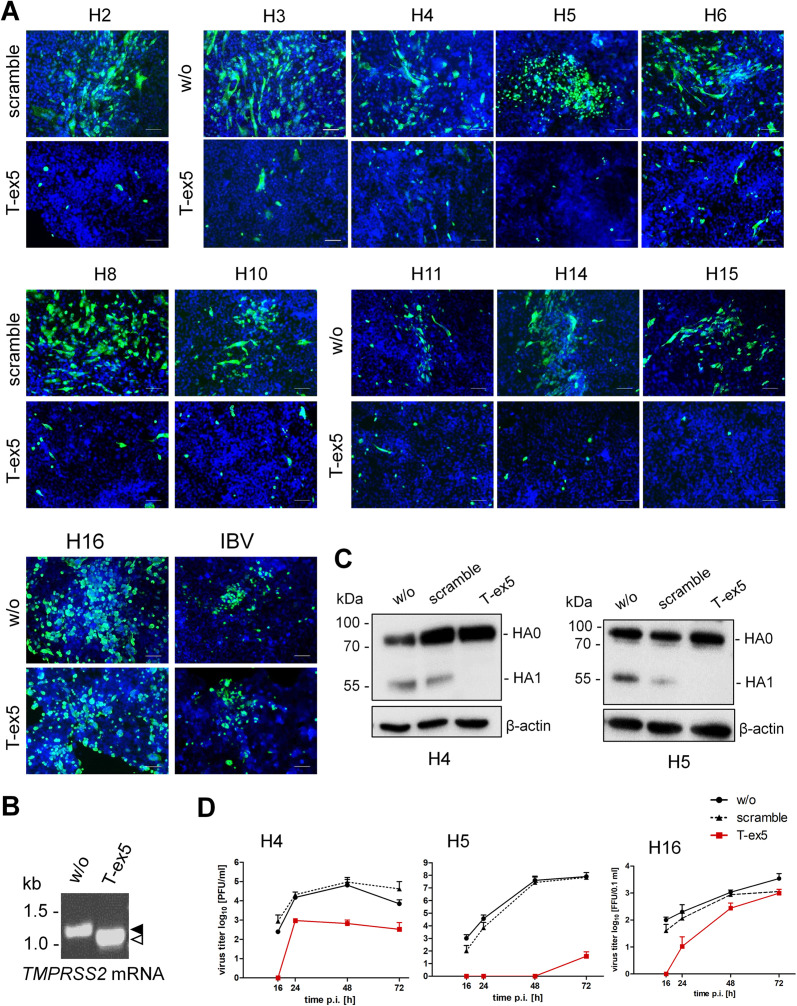
To investigate the role of TMPRSS2 in proteolytic activation of different avian HA subtypes in human airway epithelial cells, we used two infection models: the human airway cell line Calu-3 and well-differentiated cultures of primary human bronchial epithelial cells (HBEC) grown under air-liquid interface conditions. Knockdown of TMPRSS2 activity in human airway cells was performed using a previously developed TMPRSS2-specific PPMO (T-ex5). T-ex5 interferes with correct splicing of TMPRSS2 pre-mRNA, resulting in the production of TMPRSS2-mRNA, lacking exon 5, and consequently expression of a truncated TMPRSS2 variant that is enzymatically inactive (28). Recently, we demonstrated that T-ex5 PPMO supports efficient knockdown of TMPRSS2 activity in Calu-3 cells and HBEC cultures without affecting cell viability and thereby prevents proteolytic activation of human H1N1pdm and H3N2 viruses and of zoonotic H7N9 virus in these cells (22).
Calu-3 cell monolayers were treated with T-ex5 PPMO for 24 h prior to virus infection to deplete enzymatically active TMPRSS2 present in the cells. Treatment of Calu-3 cells with a PPMO of nonsense sequence (scramble) or untreated Calu-3 cells were used as controls. The cells were then infected with recombinant viruses at a low MOI and incubated without further PPMO treatment for 24 h, at which time they were fixed and immunostained against viral NP. As shown in Fig. 2A, multicycle replication and foci of infection were observed for all viruses in untreated or scramble PPMO-treated Calu-3 cells. In contrast, spread of H2, H4, H5, H6, H8, H10, H11, H14, and H15 reassortants was prevented in T-ex5 PPMO-treated cells, and only single, most likely inoculum-infected cells were visible. Inhibition of viral spread was also observed for H3-SC35M bearing an avian H3. In contrast, spread of IBV that was used as a control for TMPRSS2-independent multiplication in Calu-3 cells was similar in untreated and T-ex5-treated cells (Fig. 2A). H16-SC35M was able to replicate in both T-ex5-treated and control cells, indicating that activation of H16 is independent of TMPRSS2 in Calu-3 cells. Together, the data suggest that TMPRSS2 is crucial for proteolytic activation of all tested HA subtypes, having a monobasic cleavage site, except H16, in Calu-3 cells.
FIG 2.
Proteolytic activation of IAVs of different HA subtypes in human Calu-3 airway cells. (A) Multicycle replication of IAV and IBV in PPMO-treated Calu-3 cells. Cells were treated with 25 μM T-ex5 or scramble PPMO for 24 h or remained untreated (w/o). Cells were then inoculated with viruses at an MOI of 0.01 to 0.001 and incubated in the absence of PPMO for 24 h. Cells were fixed and immunostained for NP (green). The nuclei were stained using DAPI (blue). Representative images from two or three independent experiments are shown. Scale bars indicate 100 μm. (B) RT-PCR analysis of TMPRSS2 mRNA in PPMO-treated Calu-3 cells. Cells were treated with T-ex5 PPMO (25 μM) or remained untreated (w/o) for 24 h, the medium was replaced, and the cells were incubated in the absence of T-ex5 for 24 h. Total RNA was isolated and amplified with TMPRSS2-specific primers to amplify a full-length (filled arrowhead) and truncated Δex5 (open arrowhead) mRNA fragment. (C) Analysis of HA cleavage in PPMO-treated Calu-3 cells. Cells were treated with PPMO for 24 h as described above, inoculated with virus at an MOI of 1, and incubated for 24 h. Cell lysates were subjected to SDS-PAGE and Western blotting with HA-specific antibodies. Beta-actin served as a loading control. HA2 was not detected by H4- and H5-specific antibodies. (D) Virus growth kinetics of H4, H5, and H16 IAV in PPMO-treated Calu-3 cells. Calu-3 monolayers were treated with PPMO or remained untreated for 24 h as described above and then inoculated with H4, H5, or H16 IAV at an MOI of 0.01 to 0.001 and incubated without PPMO for 72 h. At the indicated time points, virus titers were determined by plaque assay or focus-forming assay (H16-SC35M). Data are mean values ± SD from two or three independent experiments.
Knockdown of enzymatically active TMPRSS2 expression was confirmed by analysis of TMPRSS2-mRNA in T-ex5-treated and untreated Calu-3 cells as described previously (22, 28). Only truncated TMPRSS2-mRNA lacking exon 5 was present in T-ex5-treated Calu-3 cells, indicating efficient knockdown of TMPRSS2 activity, whereas full-length TMPRSS2-mRNA was present in untreated control cells (Fig. 2B).
To examine HA cleavage in PPMO-treated Calu-3 cells, cells were incubated with T-ex5 or scramble PPMO for 24 h or remained untreated and were then inoculated with either H4-SC35M or H5N1-PR8 at an MOI of 1. At 24 h p.i., cell lysates were subjected to SDS-PAGE under reducing conditions and Western blot analysis using HA-specific antibodies. Cleavage of HA0 was observed for H4 and H5 in untreated and scramble PPMO-treated cells. In contrast, only noncleaved H4 and H5 HA0 was detected in Calu-3 cells treated with T-ex5 PPMO (Fig. 2C). The data confirm that inhibition of virus multicycle replication in T-ex5-treated cells is caused by inhibition of HA cleavage and, thus, generation of noninfectious progeny virus in the cells.
Finally, to analyze virus multicycle replication in the absence and presence of active TMPRSS2 in Calu-3 cells more quantitatively, virus growth kinetics of H4-SC35M, H5N1-PR8, and H16-SC35M as representative viruses for TMPRSS2-dependent versus TMPRSS2-independent HA activation were determined in PPMO-treated cells. To this end, Calu-3 monolayers were treated with 25 μM T-ex5 or scramble PPMO or remained untreated for 24 h as described above. The cells were then inoculated with virus at an MOI of 0.01 to 0.001 and incubated without further PPMO treatment for 72 h. Virus multiplication was determined by plaque assay (H4-SC35M and H5N1-PR8) or by focus formation assay (H16-SC35M) of supernatants collected at different times p.i. Multicycle replication of H4-SC35M was strongly inhibited in T-ex5-treated cells, and replication of H5N1-PR8 was almost completely blocked, whereas efficient replication of both viruses was observed in untreated and scramble PPMO-treated control cells (Fig. 2D). The data indicate that TMPRSS2 is crucial for activation of H4 and H5 HA with a monobasic cleavage site in Calu-3 cells. Multiplication of H16-SC35M was delayed and strongly reduced at 16 and 24 h p.i. in T-ex5-treated Calu-3 cells, but the virus was still able to undergo multicycle replication, resulting in an 8-fold final reduction in virus titer at 72 h compared to control cells. The data suggest that TMPRSS2 supports cleavage of H16 HA in Calu-3 cells but that at least one other protease is involved in H16 HA cleavage in these cells.
Together, the results demonstrate that TMPRSS2 is crucial for proteolytic activation of avian IAV HA of subtypes H2, H3, H4, H5, H6, H8, H10, H11, H14, and H15 in Calu-3 human airway epithelial cells, similar to what was previously reported for human H1, H3, and zoonotic H7 with monobasic cleavage sites (22). Conversely, activation of H16 HA was found to be largely independent of TMPRSS2 activity in Calu-3 cells.
H9N2 IAVs vary in their TMPRSS2 dependency in Calu-3 cells.
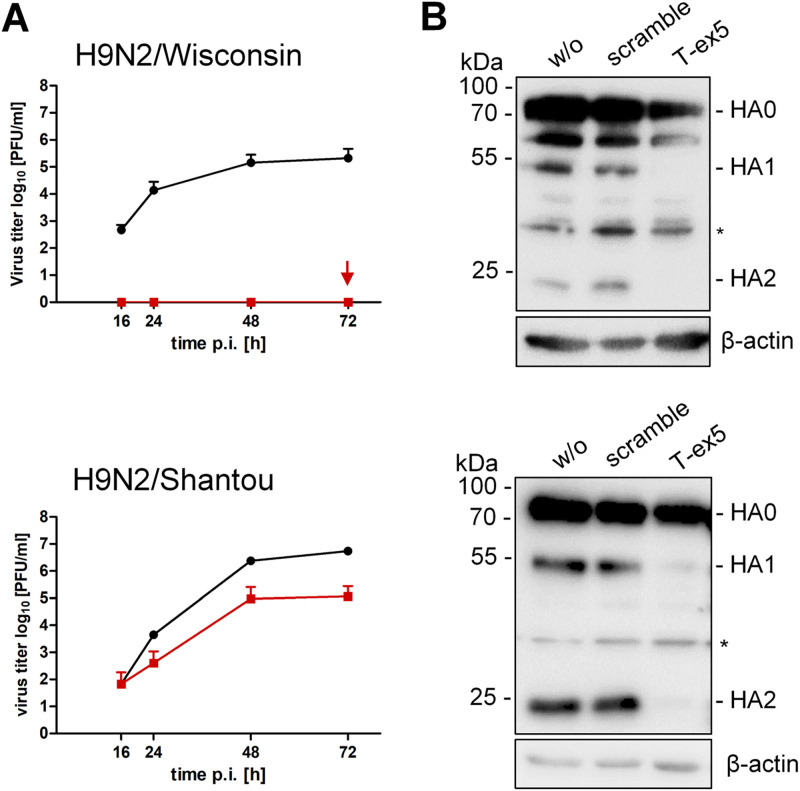
Among LPAIV subtypes, H9N2 viruses vary remarkably in amino acid sequence at the cleavage site. While H9N2 IAVs endemic to America, Europe, and Africa contain diverse monobasic cleavage site motifs, many H9N2 viruses from Asia and the Middle East possess the di- or tribasic cleavage site motif R-S-S-R↓ or R-S-R-R↓. In a previous study, we demonstrated that H9 IAVs possessing an R-S-S/R-R↓ cleavage site motif were activated by the TMPRSS2-related protease matriptase (also designated ST14) in MDCK cells and in primary chicken kidney epithelial cells in vitro, whereas H9 IAV with monobasic cleavage site motif V-S-S-R↓ was not (29).
We therefore examined the role of TMPRSS2 in proteolytic activation of two H9N2 viruses that differ in their cleavage site motif in Calu-3 cells. H9N2/Shantou possesses the cleavage site sequence R-S-S-R↓, whereas H9N2/Wisconsin contains the monobasic motif V-S-S-R↓. Calu-3 monolayers were treated with 25 μM T-ex5 PPMO or remained untreated for 24 h. Cells were then infected with H9N2/Shantou or H9N2/Wisconsin at a low MOI and incubated for 72 h. At the indicated time points, virus titers were determined by plaque assay. Multicycle replication of H9N2/Wisconsin was completely blocked in T-ex5-treated cells, while efficient virus replication was observed in control cells (Fig. 3A). In contrast, virus growth of H9N2/Shantou was markedly reduced in T-ex5-treated cells but still substantial, with a 10- to 100-fold reduction in virus titers compared to untreated cells. To examine HA cleavage of both viruses in the absence and presence of TMPRSS2 activity in Calu-3 cells, cells were treated with T-ex5 or scramble PPMO for 24 h or left untreated. Cells were then inoculated with virus at an MOI of 1, and cell lysates were subjected to SDS-PAGE and Western blotting at 24 h (H9N2/Wisconsin) or 48 h (H9N2/Shantou) p.i. Cleavage of HA0 into HA1 and HA2 was detected for both viruses in untreated and scramble PPMO-treated cells (Fig. 3B). In T-ex5-treated cells, cleavage of HA0 of H9N2/Shantou was strongly suppressed, and only small amounts of HA1 and HA2 were detected. HA cleavage of H9N2/Wisconsin was completely inhibited in T-ex5-treated cells, consistent with the observed inhibition of virus replication in these cells. The data show that H9 HA with an R-S-S-R↓ but not a V-S-S-R↓ cleavage site can be activated by protease(s) in addition to TMPRSS2 in Calu-3 cells, although less efficiently than by TMPRSS2.
FIG 3.
Proteolytic activation of H9N2 IAV in Calu-3 cells. (A) Calu-3 cells were treated with T-ex5 PPMO (25 μM) or remained untreated (w/o) for 24 h and then were inoculated with H9N2/Shantou or H9N2/Wisconsin at a low MOI and incubated without further PPMO treatment for 72 h. Supernatants were collected at 16, 24, 48, and 72 h p.i., and viral titers were determined by plaque assay. Replication of H9N2/Wisconsin was completely blocked in T-ex5-treated cells (indicated by an arrow). Data are mean values ± SD from three independent experiments. (B) HA cleavage analysis. Calu-3 cells were treated with PPMO for 24 h as described above and then inoculated with H9N2/Shantou or H9N2/Wisconsin at an MOI of 1. At 24 h (H9N2/Wisconsin) or 48 h (H9N2/Shantou) p.i., cell lysates were subjected to SDS-PAGE and Western blotting using an H9N2-specific antiserum. Unspecific protein bands are indicated by asterisks. Beta-actin served as a loading control.
Role of TMPRSS2 in proteolytic activation of HAs in HBEC.
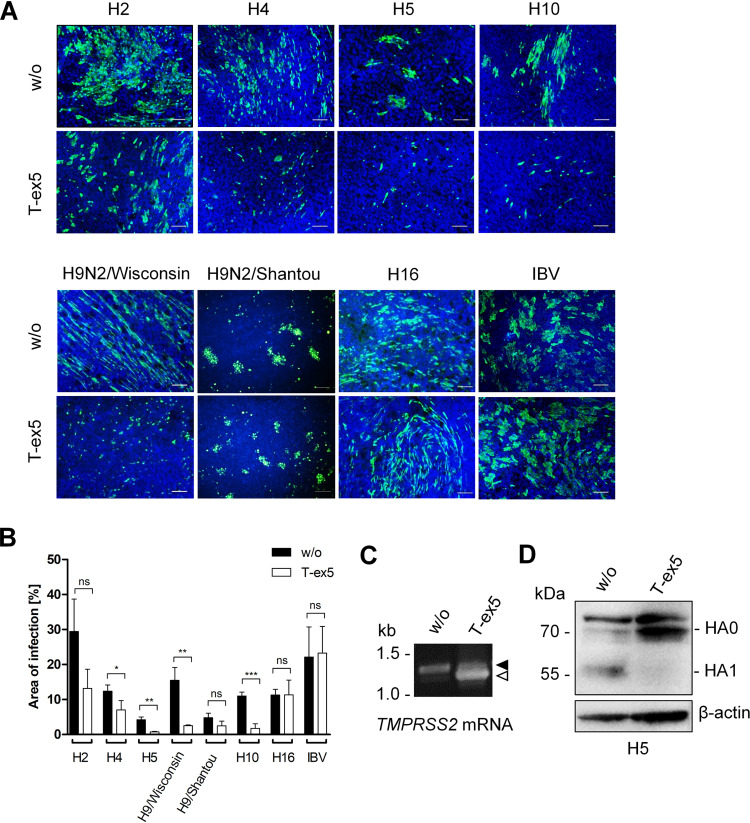
Recent studies by us and others have shown that primary HBEC express a wider array of proteases than Calu-3 cells, including HAT/TMPRSS11D and TMPRSS13/MSPL, capable of cleaving HA with monobasic cleavage sites (22, 30). Therefore, we analyzed whether TMPRSS2 is also crucial for activation of different HA subtypes in primary HBEC or whether additional proteases are sufficient to support HA cleavage in these cells. HBEC were grown under air-liquid interface conditions for 3 to 4 weeks to form a pseudostratified epithelium containing ciliated as well as mucous-producing cells that closely resembles human respiratory epithelium. Due to the limited availability of HBEC, only a subset of HA reassortants was analyzed for multicycle replication with or without PPMO-induced knockdown of TMPRSS2 activity. An overview of the HA subtypes analyzed for proteolytic activation in the different cell cultures in the absence or presence of TMPRSS2 is shown in Table 2. HBEC cultures were treated with PPMO for 24 h, inoculated at a low MOI, incubated for 24 h, and then fixed and immunostained for viral NP. Multicycle replication and spread were observed for all viruses in untreated control cells (Fig. 4A). In contrast, foci of infection of H2, H4, H5, and H10 reassortants and H9N2/Wisconsin were strongly inhibited in T-ex5 PPMO-treated HBEC, suggesting that TMPRSS2 is crucial for activation of H2, H4, H5, H9, and H10 with a monobasic cleavage site in primary HBEC. In contrast, multicycle replication and spread of H9N2/Shantou and H16-SC35M were visible in both T-ex5-treated cells and control cells, demonstrating that cleavage of H9 with the R-S-S-R↓ motif and H16 is independent of TMPRSS2 in HBEC, similar to that observed in Calu-3 cells. Virus spread in T-ex5-treated HBEC and control cells were quantified by image-based quantification using Fiji (Fig. 4B). This analysis confirmed the significant reduction of virus focus areas in T-ex5-treated versus untreated HBEC cultures infected with reassortant viruses bearing H2, H4, H5, H10, and H9N2/Wisconsin. In contrast, no significant decrease in virus spread was observed in T-ex5 PPMO-treated HBEC infected with H9N2/Shantou, H16-SC35M, or IBV. We note, however, that the focus area of H9N2/Shantou in T-ex5-treated cells was reduced compared to that in control cells. Reverse transcription-PCR (RT-PCR) analysis of PPMO-treated and untreated HBEC revealed that the majority of TMPRSS2-mRNA present in T-ex5-treated cells was truncated due to the T-ex5-induced deletion of exon 5, indicating efficient knockdown of TMPRSS2 activity in the cells (Fig. 4C). To analyze HA cleavage in HBEC with and without T-ex5 treatment, HBEC cultures were treated with PPMO for 24 h as described above and then inoculated with H5N1-PR8 at an MOI of 0.5 and incubated without PPMO for a further 72 h. Cell lysates were analyzed by SDS-PAGE and Western blotting using H5-specific antibodies. HA0 of H5N1-PR8 was efficiently cleaved in untreated cells, whereas no HA0 cleavage was detected in T-ex5 PPMO-treated cells (Fig. 4D).
TABLE 2.
Overview of experimental analyses performed with viruses carrying different HA subtypes in this studya
| Virus | Mouse lung cells |
Calu-3 cells |
HBEC |
Duck TMPRSS2 |
||||
|---|---|---|---|---|---|---|---|---|
| Virus growth | WB | Virus growth | WB | Virus growth | WB | Virus growth | WB | |
| H1-SC35M | ✓ | ND | ND | ND | ND | ND | ✓ | ND |
| H2-SC35M | ✓ | ND | ✓ | ND | ✓ | ND | ✓ | ND |
| H3-SC35M | ✓ | ND | ✓ | ND | ND | ND | ND | ND |
| H4-SC35M | ✓ | ✓ | ✓ | ✓ | ✓ | ND | ✓ | ✓ |
| H5N1-PR8 | ✓ | ND | ✓ | ✓ | ✓ | ✓ | ✓ | ND |
| H6-SC35M | ✓ | ND | ✓ | ND | ND | ND | ✓ | ND |
| H7-PR8 | NDb | NDb | NDb | NDb | NDb | NDb | ✓ | ✓ |
| H8-SC35M | ✓ | ND | ✓ | ND | ND | ND | ✓ | ND |
| H9-SC35M | ✓ | ND | ND | ND | ND | ND | ✓ | ND |
| H10-SC35M | ✓ | ND | ✓ | ND | ✓ | ND | ✓ | ND |
| H11-SC35M | ✓ | ND | ✓ | ND | ND | ND | ✓ | ND |
| H14-SC35M | ✓ | ND | ✓ | ND | ND | ND | ✓ | ND |
| H15-SC35M | ✓ | ND | ✓ | ND | ND | ND | ✓ | ND |
| H16-SC35M | ✓ | ND | ✓ | ND | ✓ | ND | ✓ | ND |
| H3N2/Victoria | ✓ | ND | ND | ND | ND | ND | ND | ND |
| H3N2/HongKong | ND | ND | ND | ND | ND | ND | ✓ | ✓ |
| H9N2/Shantou | ND | ND | ✓ | ✓ | ✓ | ND | ND | ND |
| H9N2/Wisconsin | ND | ND | ✓ | ✓ | ✓ | ND | ND | ND |
| IBV | ✓ | ✓ | ✓ | NDb | ✓ | NDb | ND | ND |
HA reassortants or wild-type viruses were analyzed for multicycle replication (virus growth) and HA cleavage by Western blotting (WB) in the indicated cell cultures and in MDCK cells transiently expressing duck TMPRSS2. WB analysis of HA cleavage was limited to HA subtypes H3, H4, H5, H7, H9, and IBV HA due to the availability of HA-specific antibodies. ✓, analysis performed; ND, not determined.
Determined in Limburg et al. (22).
FIG 4.
Proteolytic activation of IAVs of different HA subtypes in HBEC. (A) Multicycle replication of IAV and IBV in T-ex5 PPMO-treated HBEC. Well-differentiated HBEC were treated with 25 μM T-ex5 for 24 h or remained untreated (w/o). Cells were then inoculated with the indicated viruses at an MOI of 0.1 to 0.5 and incubated in the absence of PPMO for 24 h. Cells were fixed, permeabilized, and immunostained for viral NP (green). The nuclei were counterstained using DAPI (blue). Representative images from two or three independent experiments are shown. Scale bars indicate 100 μm. (B) Quantitative analysis of virus infection in PPMO-treated HBEC. Immunofluorescence data of three to four representative images of multicycle replication in T-ex5-treated and untreated HBEC were segmented, and the ratio of NP-stained area to total area was measured. Data are mean values ± SD. Statistical analysis was performed using two-sample t test. P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***) were considered significant, whereas P values of >0.05 (ns) were not. (C) RT-PCR analysis of TMPRSS2-mRNA in HBEC with or without T-ex5 treatment for 24 h. Total RNA was isolated and amplified with TMPRSS2-specific primers to amplify a full-length mature mRNA fragment (filled arrowhead) and truncated Δex5 fragment (open arrowhead), respectively. (D) Analysis of H5 HA cleavage in T-ex5 PPMO-treated HBEC. HBEC cultures were treated with T-ex5 PPMO as described above for 24 h or remained untreated and then inoculated with H5N1-PR8 at an MOI of 0.5. At 72 h p.i., cell lysates were subjected to SDS-PAGE and Western blotting using H5-specific antibodies. HA2 was not detected by the antibody. Beta-actin served as a loading control.
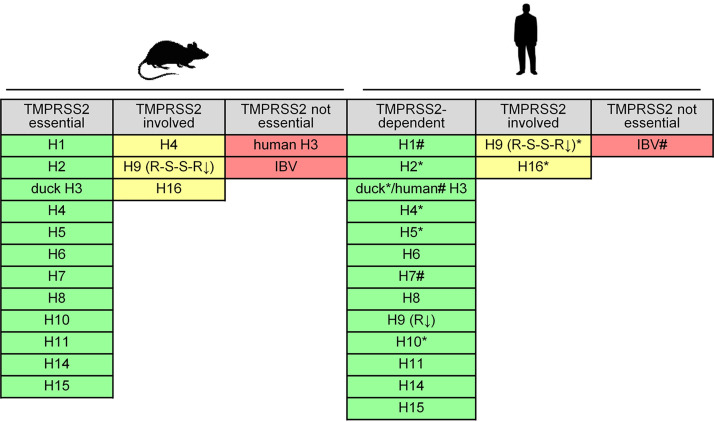
In summary, our data demonstrate that TMPRSS2 is crucial for proteolytic activation of avian IAVs of HA subtypes H1, H2, H3, H4, H5, H6, H8, H9, H10, H11, H14, and H15, having a monobasic cleavage site in respiratory cells of human and mouse. Only activation of H9 HA with R-S-S-R↓ cleavage site motif and H16 HA was supported by proteases in addition to TMPRSS2, although proteolytic activation was more efficient in TMPRSS2-expressing cells. The data are summarized in Fig. 5.
FIG 5.
Role of TMPRSS2 in activation of IAV with H1 to H16 HAs and IBV in primary murine lung cells and Calu-3 human airway cells. Results were validated in HBEC cultures in the present study (*) and in reference 22 (#).
Activation of IAV HAs by a TMPRSS2-orthologous protease from duck.
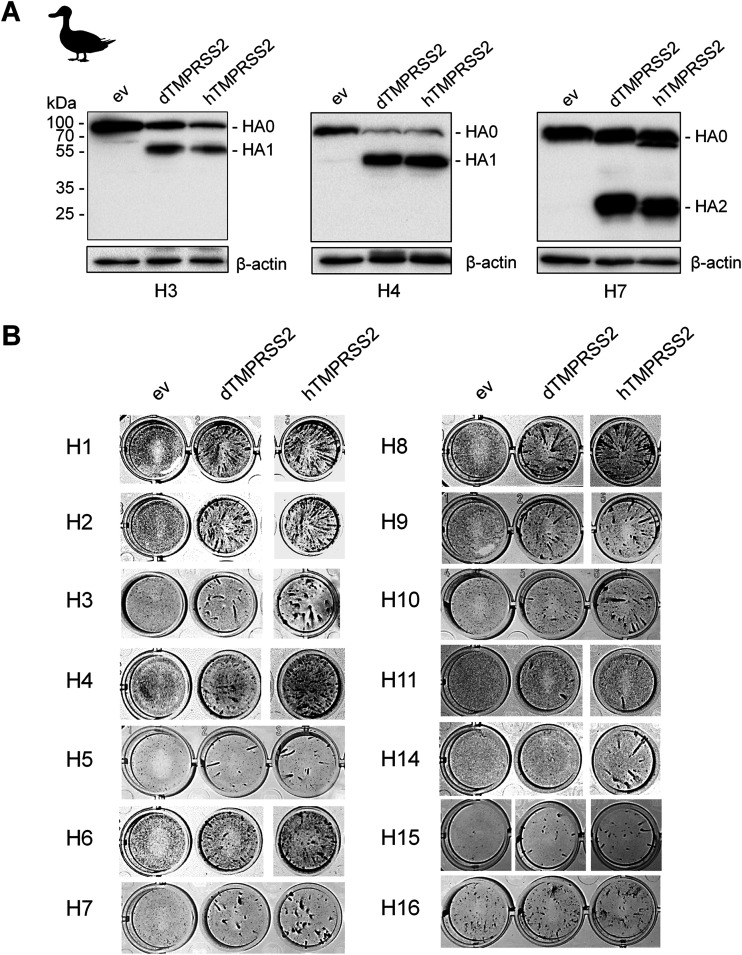
A gene encoding a TMPRSS2-orthologous protease is found in the genome of diverse avian species. Duck and chicken TMPRSS2 orthologs share 76.99% and 80.5% amino acid identity with human TMPRSS2, respectively. To examine whether TMPRSS2 from duck (dTMPRSS2) is able to activate influenza virus HA with a monobasic cleavage site, we cloned the dTMPRSS2 cDNA from lung tissue of mallard duck into the mammalian expression vector pCAGGS. To analyze HA cleavage by dTMPRSS2, HEK293 cells were cotransfected with plasmids encoding dTMPRSS2 and HA of pandemic virus A/HongKong/1/68 (H3N2), H4, and H7 HAs (Table 1). Coexpression of HA and human TMPRSS2 (hTMPRSS2) was used as a control. At 48 h posttransfection, cell lysates were analyzed by SDS-PAGE and Western blotting using HA-specific antibodies. Cleavage of H3, H4, and H7 HA0 was observed upon coexpression of dTMPRSS2 and hTMPRSS2, whereas no cleavage of HA0 was detected in cells transfected with empty vector (Fig. 6A). To examine dTMPRSS2-supported proteolytic activation of HA, MDCK cells were transiently transfected with pCAGGS encoding either dTMPRSS2, hTMPRSS2, or empty pCAGGS vector. The cells were then inoculated with the H1 to H11 and H14 to H16 reassortants and incubated for 24 h to allow multiple cycles of viral replication. Cells were fixed and immunostained for IAV NP. As shown in Fig. 6B, dTMPRSS2 facilitated multicycle replication of H3N2/HongKong, H4-SC35M, and H7-PR8 as well as reassortant viruses bearing H1, H2, H5, H6, H8, H9 (R-S-S-R↓), H10, H11, H15, and H16 HA in MDCK cells. The data demonstrate that cleavage of the HAs by dTMPRSS2 supports their proteolytic activation. We note that H16-SC35M was able to undergo multicycle replication in MDCK(H) cells transfected with empty vector, indicating that H16 HA is cleaved by an endogenous protease in the cells. However, virus foci were larger in diameter in dTMPRSS2- and hTMPRSS2-expressing cells, indicating enhanced activation of H16 HA in these cells due to transient expression of the proteases. Interestingly, H14 IAV was activated in MDCK cells by hTMPRSS2 but not by dTMPRSS2. Thus, our data show that dTMPRSS2 is capable of activating HAs of different subtypes except H14 HA at a monobasic cleavage site.
FIG 6.
Proteolytic activation of different IAV HA subtypes by duck TMPRSS2 (dTMPRSS2). (A) Analysis of HA cleavage by dTMPRSS2. HEK293 cells were cotransfected with pCAGGS plasmids encoding H3/HongKong, H4, or H7 HA and either dTMPRSS2 or human TMPRSS2 (hTMPRSS2). Transfection with empty vector (ev) was used as a control. After 48 h of incubation, cell lysates were subjected to SDS-PAGE and Western blotting using an HA-specific antibody. (B) Proteolytic activation and multicycle replication analysis. MDCK(H) cells were transfected with protease-encoding plasmids or empty vector (ev) as a negative control for 24 h and then inoculated with either reassortant bearing the indicated HA or human virus H3N2/HongKong at an MOI of 0.0005 to 0.01. At 24 h p.i., cells were fixed and immunostained using antibodies against NP, peroxidase-conjugated secondary antibodies, and a peroxidase substrate.
In conclusion, our data suggest that, in human and murine airway cells, TMPRSS2 is the major activating protease of IAV of almost all HA subtypes having a monobasic cleavage site. TMPRSS2-independent activation seems to be specific only to certain human H3N2 viruses in mice and to H9 HA with the R-S-S-R↓ cleavage site, H16, and IBV HA in both human and murine respiratory cells. Moreover, our data demonstrate that duck TMPRSS2 can activate HA with a monobasic cleavage site and, thus, represents a potential IAV-activating protease in waterfowl.
DISCUSSION
For a long time, the identity of which trypsin-like protease(s) cleaves IAV HA with a monobasic cleavage site in the mammalian respiratory tract has remained unclear. Previous studies by us and others identified TMPRSS2 as a host cell factor essential for activation and spread of H1, H2, H7, and H10 IAVs with a monobasic cleavage site in murine airways and demonstrated that lack of TMPRSS2 expression prevents virus replication and pathogenesis from these influenza viruses in mice. Recently, we identified TMPRSS2 as the major activating protease of human H1 and H3 and zoonotic H7N9 viruses with a monobasic cleavage site in human airway cells (22). In this study, we broaden those previous findings and demonstrate that TMPRSS2 is the major activating protease of IAVs, with HA subtypes H1 to H11, H14, and H15, with a monobasic cleavage site in human and mouse airway cells. TMPRSS2-independent activation seems to be a unique property of certain human H3N2 viruses, H9 HA with the R-S-S-R↓ cleavage site motif, H16 HA, and IBV HA, and, therefore, is an exception rather than the rule.
IAV HAs of subtypes H12 and H13 were not available for our study; thus, the role of TMPRSS2 in activation of those two HA subtypes remains to be determined. Galloway and coworkers analyzed activation of H1 to H14 and H16 HA by TMPRSS2 and the related protease human airway tryptase (HAT)/TMPRSS11D upon coexpression in vitro (31). They showed that TMPRSS2 activates H13 but not H12, while the opposite was observed for HAT and pancreatic trypsin. Thus, it remains of interest to determine the role of TMPRSS2 in activation of H12.
The amino acid sequence at the HA cleavage site is highly conserved within each IAV subtype but differs between HA subtypes (Table 1). In 1998, Chen et al. solved the H3 HA0 crystal structure and demonstrated that the cleavage site is located in a prominent surface loop (32). To date, the HA0 structure has been solved for two more subtypes by X-ray crystallography, namely, for the H1 HA of the 1918 pandemic IAV and for the H16 HA (26, 33). The H1 HA0 cleavage site loop is less exposed than H3 HA0. H16 HA0 contains an unusual α-helix structure in the cleavage site loop, with the arginine residue being obscured by the helix. H16 has been shown to be resistant to cleavage by trypsin yet to be cleavable by TMPRSS2 in vitro (26, 31) and by an undefined protease(s) in differentiated human airway cell cultures (34). Interestingly, here, H16 was one of the two IAV HA subtypes that can be activated by other proteases in addition to TMPRSS2 in murine and human airway cells. Further, H16-SC35M was proteolytically activated by an endogenous protease in MDCK(H) cells. Thus, although the H16 HA cleavage site appears to be less exposed, it seems to be accessible to a greater number of proteases than most other HA subtypes. Notably, carbohydrate side chains or amino acid sequences in close proximity to the cleavage site also can affect its accessibility and thereby may contribute to the sensitivity of different HAs to host proteases (20, 35, 36). Which protease(s) facilitates activation of H16 in human and murine airway cells and whether this protease(s) is also involved in activation of IBV HA and/or H9 with the R-S-S-R↓ cleavage site in airway cells remains to be determined. Furthermore, it is still uncertain whether the same protease(s) supports H16 HA activation in MDCK(H) cells. H16 IAV has been characterized as gull specific and is rarely isolated from ducks (37, 38). It remains to be analyzed whether activation of H16 HA by a broader range of proteases plays a role in proteolytic activation and tissue tropism of H16 IAV in gulls.
Furthermore, we demonstrated that H9 HA with the R-S-S-R↓ cleavage site motif can be activated by proteases in addition to TMPRSS2 in human and murine airway cells. Previous work by us strongly suggests that this is due to the type II transmembrane protease matriptase (29). Orthologs of matriptase are encoded in the genomes of mouse and chicken (39). Recombinant matriptase cleaved H9 of H9N2/Shantou (R-S-S-R↓) but not HA of H9N2/Wisconsin (V-S-S-R↓) (29). Moreover, H9N2/Shantou was able to replicate in primary chicken embryo kidney (CEK) cells in the absence of exogenous trypsin, whereas H9N2/Wisconsin was not. H9N2/Shantou multicycle replication in CEK was inhibited by a peptide mimetic inhibitor of matriptase. Because matriptase is expressed in a broad range of tissues, it may affect tissue tropism of H9N2 IAV (39, 40). For example, the nephrotropism observed with H9N2 viruses in poultry might be due to virus activation by matriptase in kidney cells. H9N2 viruses in domestic poultry have become endemic in several geographical areas since the mid-1990s and are occasionally transmitted to humans, raising concern about their pandemic potential (6, 8). Importantly, H9N2 viruses have contributed internal genes to H5N1, H5N6, H7N9, and H10N8 viruses that can infect humans (reviewed in reference 41). Here, however, activation of H9 HA with the R-S-S-R↓ cleavage site by putative matriptase was less efficient than H9 HA activation by TMPRSS2 in both human and murine cells. Interestingly, TMPRSS2 has been shown to activate matriptase in prostate cells (42). Thus, knockdown/knockout of TMPRSS2 activity also might reduce matriptase activity in airway cells. It remains to be examined whether the lower level of H9 activation in cells with knockdown/knockout of TMPRSS2 activity is a matter of reduced efficiency of matriptase in H9 HA cleavage compared to TMPRSS2 or instead due to decreased levels of matriptase activity.
Here, we found that H4 HA is proteolytically activated in lung explants and AECII of Tmprss2−/− mice, although the levels of HA cleavage and virus multiplication were very low, and it remains to be investigated whether H4-SC35M is able to replicate efficiently in Tmprss2−/− mice in vivo. Interestingly, H4 HA contains the cleavage site motif K-A-S-R↓, possessing a lysine in the P4 position. Matriptase shows a preference for basic residues in both P4 and P1 positions and, thus, might contribute to H4 HA cleavage in mice. However, H3 HA also possesses K in P4 position (K-Q-T-R↓), and previous studies by us and others showed that H3 HA is not cleaved by matriptase (29, 43, 44). Thus, it remains to be investigated whether matriptase contributes to H4 HA cleavage in mice. Importantly, H4-SC35M was not able to undergo multicycle replication in Calu-3 cells or HBEC with T-ex5 PPMO-induced knockdown of TMPRSS2 activity. Thus, activation of H4 HA in human airway cells is TMPRSS2 dependent, in contrast to the activation scenario in mice.
Previous studies have revealed that human and mouse can differ in their HA-cleaving protease repertoire (19, 20, 27). Certain human H3N2 viruses replicate independently of TMPRSS2 in mice yet are dependent on TMPRSS2 for HA activation in human cells. Activation of IBV HA was shown to be TMPRSS2 independent in murine AECII, whereas TMPRSS2 was necessary for IBV activation in human AECII (21, 22). Thus, previous studies suggest that activation of H3 in mice is facilitated by a mouse-specific protease or a protease not present as an active enzyme in human airways (22, 27). Notably, the human genome encodes 588 proteases, whereas 672 protease genes have been annotated in the murine genome (45). Moreover, differences in the substrate specificity of murine proteases and their human orthologues might contribute to the protease repertoire available for HA cleavage. Correspondingly, the murine proteases prostasin and hepsin have been shown to cleave H3 HA in vitro, whereas their human orthologues did not (27). On the contrary, human but not murine kallikrein (KLK5) was demonstrated to be able to cleave H3 HA in vitro (46). Finally, different protease inhibitor repertoires available might lead to differences in HA activation between human and mouse. A number of physiological protease inhibitors, including plasminogen-activator inhibitor 1 (SERPIN1), hepatocyte growth factor activator inhibitor 2 (HAI-2), or serine peptidase inhibitor, Kunitz type 1 (SPINT1, also designated HAI-1), have been demonstrated to inhibit IAV activation in cell culture and in mouse models (47–49). Therefore, it may be interesting to compare the distribution of these inhibitors in airway tissues of human versus mouse as well as their expression along the human respiratory tract. Activation of IBV was shown to be independent of TMPRSS2 in Calu-3 cells and primary HBEC but was strictly TMPRSS2 dependent in primary human AECII, suggesting that AECII either lack expression of an appropriate protease or that the protease is inhibited by a cognate inhibitor.
The proteases that support activation of IAV in waterfowl and poultry remain poorly defined. Comparative analysis of zebra finch, chicken, human, and mouse genomes indicates that avian genomes encode a considerably smaller number of proteases than mammalian genomes (50). Here, we demonstrate that TMPRSS2 from duck is able to activate almost all HA subtypes in vitro, suggesting that TMPRSS2 supports HA activation in ducks. Furthermore, TMPRSS2 from chicken was shown to cleave the HA of the 1918 (H1N1) pandemic virus in vitro, indicating that TMPRSS2 supports IAV activation in chicken as well (51). Interestingly, H14 HA was cleaved by human TMPRSS2 but not by duck TMPRSS2. H14 HA contains a lysine instead of an arginine in position P1 at the cleavage site (Table 1). Thus, the data indicate that TMPRSS2 orthologs from human and duck differ in their ability to cleave at a lysine residue. It has yet to be demonstrated whether TMPRSS2 plays a critical role in HA activation in ducks and chicken and whether the avian intestinal and respiratory tracts differ in their HA-cleaving protease repertoire.
It has been established that humans can be infected with avian IAV of subtypes H5, H6, H7, H9, and H10 by exposure to infected wild birds and poultry. Infections are associated with diverse symptoms such as conjunctivitis, influenza-like illness, pneumonia, respiratory distress syndrome, and encephalitis (reviewed in reference 5). Fortunately, in most cases the virus cannot transmit from human to human due to several species barriers. However, on rare occasions IAV can establish a new virus lineage in a mammalian host. Adaptation of avian IAV to humans typically requires several genetic alterations and is a complex process, but some factors were recognized as key to the process, including the receptor-binding specificity of HA and its pH stability as well as adaptation of the viral polymerase complex to facilitate nuclear import and replication of the viral genome in mammalian cells (reviewed in reference 52). Here, we demonstrate that human TMPRSS2 is crucial for activation of avian IAV HAs of various subtypes in human airway cells. Further, we show that duck TMPRSS2 activates avian HAs with a monobasic cleavage site. Thus, our data suggest that HA cleavage is a conserved mechanism that requires no adaptation for IAV in order to cross from avian to human hosts. Orthologues of TMPRSS2 from swine and nonhuman primates have been shown to be capable of cleaving HA with a monobasic cleavage site in vitro, indicating that TMPRSS2 supports HA activation in various species (53, 54). It remains to be determined, however, whether efficiency of TMPRSS2-mediated HA cleavage differs between host species and, thus, can contribute to the viral host range and interspecies transmission.
The present study provides further evidence that TMPRSS2 is an essential host cell factor for IAV replication in human airways and therefore provides a potential drug target. The concept of targeting HA activation for influenza treatment was first addressed in the 1980s by Zhirnov and coworkers using the broad-range serine protease inhibitor aprotinin from bovine lung (55, 56). Aprotinin efficiently inhibited influenza virus activation and multiplication in human airway cells in vitro and in experimentally infected mice. Furthermore, aerosolized aprotinin inhalations in patients with influenza and parainfluenza markedly reduced the duration of symptoms without causing side effects in a clinical trial (56). Nevertheless, the development of protease inhibitors for influenza treatment has been minimal to date, most likely due to limited knowledge about proteases involved and their specific physiological functions. TMPRSS2-deficient mice show no adverse phenotype compared to wild-type littermates, indicating functional redundancy or compensation of physiological functions by another protease(s). TMPRSS2 has been shown to be sensitive to a number of broad-spectrum serine protease inhibitors, including aprotinin, camostat, and nafamostat (57–60). Additionally, a number of potent peptide or peptide-mimetic inhibitors of TMPRSS2 have been described and were shown to prevent IAV and IBV activation and multiplication in cell cultures (reviewed in reference 61). However, all these compounds inhibit numerous trypsin-like proteases and therefore may cause various side effects. In contrast, specific inhibition of TMPRSS2 during an acute influenza infection is expected to be well tolerated. Unfortunately, to date no structural information on the catalytic domain of TMPRSS2 is available; thus, the development of highly selective peptide or peptide-mimetic inhibitors of TMPRSS2 is hampered. Alternatively, PPMO are highly selective inhibitors of target gene expression (62). They bind to a complementary sequence in target mRNA and can affect gene expression by steric blockage of translation initiation or pre-mRNA splicing. T-ex5 PPMO is the first highly selective inhibitor of TMPRSS2 activity that has been developed (28). Recently, we demonstrated that T-ex5 strongly inhibits replication of SARS-CoV-2 in Calu-3 cells due to inhibition of proteolytic activation of the spike protein S and thereby identified TMPRSS2 as an essential host cell factor for SARS-CoV-2 (59). TMPRSS2 has been demonstrated to activate several respiratory viruses with monobasic cleavage sites in their fusion proteins, including human and zoonotic coronaviruses (CoV), such as HCoV-229E, severe acute respiratory syndrome CoV, and Middle East respiratory syndrome CoV, human metapneumovirus, and parainfluenza viruses in vitro (reviewed in reference 63). Thus, the development of TMPRSS2-specific inhibitors may provide a treatment approach for a broad range of respiratory virus infections. Furthermore, the present study suggests that TMPRSS2 inhibitors provide useful drugs for the treatment of human infections with avian IAV and should be considered an approach for pandemic preparedness as well. Based on our findings here, further toxicity and antiviral evaluations of TMPRSS2-specific PPMO in animal models are warranted.
In conclusion, our study extends previous findings on proteolytic activation of IAV HA with monobasic cleavage sites and provides evidence that TMPRSS2 is the major activating protease of avian HAs of subtypes H1 to H11, H14, and H15 in human airway cells. Furthermore, our data indicate that TMPRSS2 supports activation of IAV in ducks.
MATERIALS AND METHODS
Cells.
Propagation of all cells was carried out at 37°C and 5% CO2. Calu-3 human airway epithelial cells (ATCC number HTB55) were cultured in Dulbecco’s modified Eagle medium (DMEM)–F-12 Ham (1:1) (Gibco) supplemented with 10% fetal calf serum (FCS), penicillin, streptomycin, and glutamine, with fresh culture medium replenished every 2 to 3 days. HEK293 human embryonic kidney cells, Madin-Darby canine kidney II (MDCKII) cells, and MDCK(H) cells were maintained in DMEM supplemented with 10% FCS, antibiotics, and glutamine.
Primary human bronchial epithelial cells (HBEC; purchased from Lonza) were cultivated under air-liquid interface conditions to form well-differentiated, pseudostratified cultures as described previously (22). Briefly, HBEC were maintained in hormone- and growth factor-supplemented airway epithelial cell growth medium (AEGM, ready-to-use; PromoCell). At 80% confluence, cells were detached with 0.05% trypsin-EDTA (Gibco) and seeded on membrane supports (12-mm Transwell culture inserts, 0.4-μm pore size; Costar) coated with 0.05 mg collagen from calf skin (Sigma-Aldrich) in ready-to-use AEGM supplemented with 1% penicillin-streptomycin. HBEC were cultured for 2 days until they reached complete confluence. The apical medium then was removed and the basal medium was replaced by a 1:1 mixture of DMEM (Sigma) and ready-to-use AEGM supplemented with 60 ng/ml retinoic acid (Sigma). Cultures were maintained under air-liquid interface conditions for 4 weeks with medium in the basal chamber replenished every 2 to 3 days. Fully differentiated 4-week-old cultures were used for the experiments.
Plasmids.
pHW2000 plasmids encoding the gene segments of SC35M were kindly provided by Thorsten Wolff (Robert Koch Institute, Berlin, Germany). pHW2000 plasmids carrying the gene segments of A/PR8/34 (H1N1) (PR8) and pHW2000 plasmids carrying HA and NA genes of A/Vietnam/1203/2004 (H5N1) were kindly provided by Richard Webby and Robert Webster (St. Jude Children's Research Hospital, Memphis, TN). pHW2000 plasmids carrying HA of A/sentinel mallard/Germany/S/Ra517K/2007 (H2N5), A/mallard/Germany/1240/1/2007 (H4N6), A/turkey/Germany/R617/2007 (H6N2), A/turkey/Ontario/6118/1968 (H8N4), A/chicken/Emirates/R66/2002 (H9N2), A/mallard/Germany/R2075/2007 (H10N7), A/domestic duck/Germany/R784/2006 (H11N1), A/mallard/Gurijev/263/1982 (H14N3), and A/shearwater/West Australia/2576/1979 (H15N9) have been described previously (24). The cDNA encoding HA of A/black-headed gull/Sweden/5/99 (H16N3; accession number AY684891.1) was purchased from ThermoFisher and was cloned into the pHW2000 vector using BsmBI restriction sites.
Virus isolates and recombinant viruses.
The wild-type IAVs used in this study were A/Victoria/5/1975 (H3N2), kindly provided by Stephan Pleschka (Institute of Virology, Gießen, Germany), A/turkey/Wisconsin/1/1966 (H9N2), A/quail/Shantou/782 (H9N2), and B/Malaysia/2506/2004. Recombinant human virus H5N1-PR8 (containing HA and NA of A/Vietnam/1203/2004 with the other 6 gene segments of A/PR8/34 [H1N1]) has been described elsewhere (25). Recombinant A/Hong Kong/1/68 (H3N2) virus was described in reference 64.
Recombinant HA-SC35M (1:7) viruses were generated by reverse genetics using the pHW2000-based eight-plasmid system described in reference 65. Briefly, HEK293 cells were cotransfected with pHW2000 plasmid carrying a representative avian HA and seven plasmids encoding the remaining gene segments of SC35M by using Lipofectamine 2000 (Invitrogen). At 48 h after transfection, the cell supernatant was treated with 1 μg/ml tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma) for 1 h at 37°C and inoculated in MDCK(II) cells, followed by 1 h of incubation. The cells were washed and maintained in infection medium containing 1 μg/ml TPCK-trypsin for 2 to 3 days. Virus-containing supernatants were analyzed by plaque assay and sequencing, and stock viruses were propagated in MDCK(II) cells as described below.
All IAVs except for H9N2/Shantou, H9N2/Wisconsin, and H16-SC35M were propagated in MDCK(II) cells in infection medium containing 1 μg/ml TPCK-treated trypsin. H9N2 viruses were grown in the allantoic cavity of 11-day-old embryonated chicken eggs. Cell supernatants and allantoic fluid were cleared by low-speed centrifugation and stored at –80°C. H16 HA is not cleaved by trypsin (26). Therefore, the generation of recombinant H16-SC35M virus was modified as follows: plasmid-transfected HEK293 cells were incubated for 72 h, the cell pellets were harvested, diluted in phosphate-buffered saline (PBS), and injected into the allantoic cavity of 11-day-old embryonated chicken eggs. After 72 h, allantoic fluid was cleared by low-speed centrifugation and stored at –80°C.
Antibodies.
Antibodies used in this study were polyclonal rabbit serum anti-H5N1 (AP301060PU-N; OriGene Technologies GmbH), anti-H4 HA (11714-RP01; SinoBiologicals), anti-H8 HA (11722-RP01; SinoBiologicals), anti-H9N2 (PA5-81658; Invitrogen), and anti-H3 HA (GTX127363; GeneTex). A monoclonal mouse anti-beta actin antibody was purchased from Abcam (ab6276). Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Dako. Immunofluorescence analyses were performed using the following antibodies: monoclonal mouse anti-IAV nucleoprotein (NP) (Abcam) and mouse anti-IBV NP (Thermo Fisher Scientific). Species-specific fluorescein isothiocyanate-conjugated and Alexa Fluor dye-conjugated secondary antibodies were purchased from Dako and Life Technologies, respectively.
PPMO.
Phosphorodiamidate morpholino oligomers (PMO) were synthesized at Gene Tools, LLC (Corvallis, OR, USA), and have been described previously (28). PMO sequences (5′ to 3′) were the following: T-ex5, CAGAGTTGGAGCACTTGCTGCCCA; scramble, CCTCTTACCTCAGTTACAATTTATA. The cell-penetrating peptide (RXR)4 (R is arginine and X is 6-aminohexanoic acid) was covalently conjugated to the 3′ end of each PMO through a noncleavable linker to produce peptide-PMO (PPMO) by methods previously described (66).
RNA isolation and RT-PCR analysis of exon skipping.
For analysis of exon skipping in TMPRSS2-mRNA in PPMO-treated Calu-3 and HBEC, the cells were incubated with indicated concentrations of PPMO or without PPMO in infection medium for the indicated time points. Total RNA was isolated using the Monarch total RNA miniprep kit (NEB) according to the manufacturer’s protocol. RT-PCR was carried out with total RNA using the one-step RT-PCR kit (Qiagen) according to the supplier’s protocol. For detection of TMPRSS2 mRNAs and to analyze exon skipping, primers TMPRSS2-108fwd (5′ CTA CGA GGT GCA TCC 3′) and TMPRSS2-1336rev (5′ CCA GAG GCC CTC CAG CGT CAC CCT GGC AA 3′), designed to amplify a full-length PCR product of 1,228 bp from control cells and a shorter PCR fragment of 1,100 bp (Δex5) from T-ex5-treated cells, were used (28). RT-PCR products were resolved on a 0.8% agarose gel stained with ethidium bromide.
Preparation of murine lung explants and AECII.
TMPRSS2-deficient mice (strain Tmprss2tm1Psn; mixed C57BL/6J-129 background) were kindly provided by Peter Nelson (Fred Hutchinson Cancer Research Center, Seattle, WA) and have been described previously (16, 67). Homozygous TMPRSS2-knockout mice (Tmprss2−/−) and wild-type littermates (Tmprss2+/+) were bred and housed in the animal facility of the Philipps-University Marburg. All protocols involving mice have been approved by the Commission on Animal Protection and Experimentation at the Philipps-University Marburg. Briefly, for lung explant preparation, Tmprss2−/− mice and wild-type littermates were sacrificed using cervical dislocation. The thorax was opened to excise the larynges, trachea, bronchi, and lungs. Lungs were separated from the remaining respiratory tract, and surrounding tissue was removed. The lobes of the lung were transferred to a 35-mm dish with 3 ml explant medium (infection medium supplemented with 1% nonessential amino acids [NEAA; Gibco]), stored on ice. Tissue samples were then taken with a 4-mm biopsy punch and maintained in a 12-well plate containing 2 ml explant medium at 37°C and 5% CO2.
The isolation of alveolar epithelial cells type II (AECII) was conducted similar to previously described procedures (22, 27). The cells then were seeded on membrane supports (12-mm Transwell Clear culture inserts, 0.4-μm pore size; Costar), coated with 0.05 mg collagen type I from calf skin (Sigma-Aldrich) per well and cultivated with DMEM–FCS for 24 h at 37°C and 5% CO2. Cells were washed and incubated under air-liquid interface conditions for 24 h prior to infection.
Multicycle replication in primary murine lung explants.
The lung punch explants from Tmprss2−/− and Tmprss2+/+ mice were inoculated with 4 × 103 PFU of the used IAV for 1 h. The inoculum was removed by careful washing, and infected explants were incubated in explant medium for 72 h. At 16, 24, 48, and 72 h p.i., virus titers were determined in number of PFU/ml by plaque assay in MDCKII cells as described previously (22).
Infection of cells and multicycle viral replication.
Infection experiments and PPMO treatment of Calu-3 cells were performed using infection medium (DMEM supplemented with 0.1% bovine serum albumin [BSA], glutamine, and antibiotics). Infection and PPMO treatment of HBEC were performed under serum-free conditions using the airway medium defined above.
For analysis of multicycle replication kinetics or virus spread in Calu-3 cells with or without T-ex5 treatment, cells were seeded in 24-well plates and grown to near confluence. Cells were incubated with 25 μM T-ex5 PPMO or scramble PPMO in infection medium for 24 h or remained untreated. After removal of preinfection medium, the cells were inoculated with virus at an MOI of 0.01 to 0.001 in infection medium for 1 h, and then washed with PBS and incubated in infection medium without PPMO for 72 h. At 16, 24, 48, and 72 h p.i., supernatants were collected and viral titers determined by plaque assay in MDCK(II) cells with Avicel overlay as described previously (22). For analysis of virus spread at 24 h p.i., Calu-3 cells were fixed with 4% paraformaldehyde (PFA) and immunostained for NP as described below. Growth kinetics of H16-SC35M were determined by focus formation assay as described elsewhere (64). Briefly, for titration, confluent MDCK(H) cells grown in 96-well plates were infected with 50 μl of serial 10-fold virus dilution in infection medium as described above. The titration was performed in duplicate. The cells were then incubated for 8 h at 37°C, fixed with 4% PFA, and immunostained for NP as described below.
To analyze HA cleavage in Calu-3 cells in the presence and absence of TMPRSS2, cells were treated with T-ex5 or scramble PPMO for 24 h as described above. Cells were then inoculated with virus at an MOI of 1 for 1 h, washed with PBS, and incubated for 24 to 48 h in fresh infection medium without PPMO. Cell lysates were subjected to SDS-PAGE and Western blotting using HA-specific antibodies.
For analysis of IAV spread in HBEC, the cells were treated from the basal site with 25 μM PPMO in airway cell medium for 24 h. Cells were washed with PBS and inoculated apically with the virus at an MOI of 0.01 to 0.001 for 1 h, washed with PBS, and incubated in fresh infection medium (basal chamber) without PPMO for 24 h. At 24 h p.i., cells were fixed with methanol-acetone (1:1) on ice and immunostained for viral NP as described below. Quantitative analysis of virus foci in PPMO-treated HBEC was performed using immunofluorescence data of three to four representative images of virus spread in T-ex5-treated and untreated HBEC. Images were segmented and NP-stained area over total area was measured using Fiji. For statistical analysis, two-sample t test was used under the assumption of normal distribution.
Immunofluorescence staining and microscopy.
To analyze virus spread in HBEC, the cells were fixed and permeabilized using methanol-acetone (1:1), blocked with 2% BSA, 5% glycerol, and 0.2% Tween 20, and incubated with primary antibodies against NP of IAV. Subsequently the cells were washed and incubated with species-specific Alexa Fluor dye-conjugated secondary antibodies. 4′,6-Diamidino-2-phenylindole (DAPI) was used as a counterstain. Cells were mounted in Fluoroshield, and images were captured using a fluorescence microscope (Zeiss Axiophot or Zeiss Axiovert 200M) and camera.
SDS-PAGE and Western blot analysis.
Cells were washed with PBS, lysed using CellLytic M cell lysis reagent (Sigma-Aldrich) for 30 min on ice, and cleared from cell debris by centrifugation. Lysates were supplemented with reducing SDS-PAGE sample buffer and heated at 95°C for 10 min. Proteins were subjected to SDS-PAGE (12% gel), transferred to a polyvinylidene difluoride (PVDF) membrane, and detected by incubation with primary antibodies and species-specific peroxidase-conjugated secondary antibodies. Proteins were visualized using the ChemiDoc XRS+ system and Image Lab software (Bio-Rad).
Cloning of pCAGGS-duckTMPRSS2.
Full-length cDNA of duck TMPRSS2 (dTMPRSS2; accession number XM_038172890) was amplified from total RNA of lung tissue of a mallard duck by RT-PCR as described above with protease-specific primers. Primer sequences are available upon request. The dTMPRSS2 cDNA was subcloned into the mammalian expression plasmid pCAGGS using EcoRI and XhoI restriction sites by the “aqua cloning” method described in reference 68. Amino acid sequence alignment of dTMPRSS2, human TMPRSS2, and chicken TMPRSS2 was performed using Serial Cloner.
Multicycle virus replication in cells transiently expressing protease.
The MDCK(H) cells were seeded in 24-well plates and transfected with protease-encoding plasmids as described above. At 24 h posttransfection, cells were infected with virus at an MOI of 0.0005 to 0.01 for 1 h. Inoculum was removed, and cells were washed and incubated for 24 h in DMEM-BSA. At 24 h postinfection, cells were immunostained for viral NP as described previously (14). Briefly, cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.3% Triton X-100 in PBS. Cells were incubated with IAV NP-specific antibodies and horseradish peroxidase-conjugated secondary antibodies and stained using the peroxidase substrate TrueBlue (KPL). Images were taken with the ChemiDoc XRS+ system and Image Lab software (Bio-Rad).
ACKNOWLEDGMENTS
We are grateful to Thorsten Wolff, Richard Webby, and Robert Webster for providing the IAV reverse genetic systems and HA and NA plasmids of H5N1 virus. We thank Peter Nelson for providing the TMPRSS2-deficient mouse model. We thank Stephan Pleschka for providing the A/Victoria/3/1975 (H3N2) virus.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation), SFB 1021 (project B07) to E.B.-F., and by the LOEWE Center DRUID (project D1) to E.B.-F.
We thank Guido Schemken and the personnel of the animal facility at the BMFZ of the Philipps-University Marburg for support in generating this work.
This article is dedicated to the memory of our dear teacher and colleague Hans-Dieter Klenk, who inspired and supported our work.
Footnotes
Supplemental material is available online only.
Contributor Information
Eva Böttcher-Friebertshäuser, Email: friebertshaeuser@staff.uni-marburg.de.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Wu Y, Wu Y, Tefsen B, Shi Y, Gao GF. 2014. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol 22:183–191. 10.1016/j.tim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoon S-W, Webby RJ, Webster RG. 2014. Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol 385:359–375. 10.1007/82_2014_396. [DOI] [PubMed] [Google Scholar]
- 3.França MS, Brown JD. 2014. Influenza pathobiology and pathogenesis in avian species. Curr Top Microbiol Immunol 385:221–242. 10.1007/82_2014_385. [DOI] [PubMed] [Google Scholar]
- 4.Swayne DE, Suarez DL, Sims LD. 2013. Influenza, p 181–218. In Swayne DE (ed), Diseases of poultry. Wiley, New York, NY. [Google Scholar]
- 5.Richard M, Herfst S. 2014. Avian influenza A viruses: from zoonosis to pandemic. Future Virol 9:513–524. 10.2217/fvl.14.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. 1999. Human infection with influenza H9N2. Lancet 354:916–917. 10.1016/S0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 7.Arzey GG, Kirkland PD, Arzey KE, Frost M, Maywood P, Conaty S, Hurt AC, Deng Y-M, Iannello P, Barr I, Dwyer DE, Ratnamohan M, McPhie K, Selleck P. 2012. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis 18:814–816. 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander DJ. 2007. An overview of the epidemiology of avian influenza. Vaccine 25:5637–5644. 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 9.Peiris JSM, de Jong MD, Guan Y. 2007. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev 20:243–267. 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik Peiris JS. 2009. Avian influenza viruses in humans. Rev Sci Tech 28:161–173. 10.20506/rst.28.1.1871. [DOI] [PubMed] [Google Scholar]
- 11.Russell CJ. 2014. Acid-induced membrane fusion by the hemagglutinin protein and its role in influenza virus biology. Curr Top Microbiol Immunol 385:93–116. 10.1007/82_2014_393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Böttcher-Friebertshäuser E, Garten W, Matrosovich M, Klenk HD. 2014. The hemagglutinin: a determinant of pathogenicity. Curr Top Microbiol Immunol 385:3–34. 10.1007/82_2014_384. [DOI] [PubMed] [Google Scholar]
- 13.Garten W, Klenk H-D. 2008. Cleavage activation of the influenza virus hemagglutinin and its role in pathogenesis, p 156–167. In Klenk H-D, Matrosovich MN, Stech J (ed), Avian influenza, vol 27. Karger, Basel, Switzerland. [Google Scholar]
- 14.Böttcher E, Matrosovich T, Beyerle M, Klenk H-D, Garten W, Matrosovich M. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol 80:9896–9898. 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatesuer B, Bertram S, Mehnert N, Bahgat MM, Nelson PS, Pöhlmann S, Pöhlman S, Schughart K. 2013. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog 9:e1003774. 10.1371/journal.ppat.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarnow C, Engels G, Arendt A, Schwalm F, Sediri H, Preuss A, Nelson PS, Garten W, Klenk H-D, Gabriel G, Böttcher-Friebertshäuser E. 2014. TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice. J Virol 88:4744–4751. 10.1128/JVI.03799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai K, Ami Y, Tahara M, Kubota T, Anraku M, Abe M, Nakajima N, Sekizuka T, Shirato K, Suzaki Y, Ainai A, Nakatsu Y, Kanou K, Nakamura K, Suzuki T, Komase K, Nobusawa E, Maenaka K, Kuroda M, Hasegawa H, Kawaoka Y, Tashiro M, Takeda M. 2014. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J Virol 88:5608–5616. 10.1128/JVI.03677-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambertz RLO, Gerhauser I, Nehlmeier I, Leist SR, Kollmus H, Pöhlmann S, Schughart K. 2019. Tmprss2 knock-out mice are resistant to H10 influenza A virus pathogenesis. J Gen Virol 100:1073–1078. 10.1099/jgv.0.001274. [DOI] [PubMed] [Google Scholar]
- 19.Lambertz RLO, Gerhauser I, Nehlmeier I, Gärtner S, Winkler M, Leist SR, Kollmus H, Pöhlmann S, Schughart K. 2020. H2 influenza A virus is not pathogenic in Tmprss2 knock-out mice. Virol J 17:56. 10.1186/s12985-020-01323-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai K, Sekizuka T, Ami Y, Nakajima N, Kitazawa M, Sato Y, Nakajima K, Anraku M, Kubota T, Komase K, Takehara K, Hasegawa H, Odagiri T, Tashiro M, Kuroda M, Takeda M. 2015. A mutant H3N2 influenza virus uses an alternative activation mechanism in TMPRSS2 knockout mice by loss of an oligosaccharide in the hemagglutinin stalk region. J Virol 89:5154–5158. 10.1128/JVI.00124-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai K, Ami Y, Nakajima N, Nakajima K, Kitazawa M, Anraku M, Takayama I, Sangsriratanakul N, Komura M, Sato Y, Asanuma H, Takashita E, Komase K, Takehara K, Tashiro M, Hasegawa H, Odagiri T, Takeda M. 2016. TMPRSS2 independency for haemagglutinin cleavage in vivo differentiates influenza B virus from influenza A virus. Sci Rep 6:29430. 10.1038/srep29430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Limburg H, Harbig A, Bestle D, Stein DA, Moulton HM, et al. 2019. TMPRSS2 is the major activating protease of influenza A virus in primary human airway cells and influenza B virus in human type II pneumocytes. J Virol 93:e00649-19. 10.1128/JVI.00649-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheiblauer H, Kendal AP, Rott R. 1995. Pathogenicity of influenza A/Seal/Mass/1/80 virus mutants for mammalian species. Arch Virol 140:341–348. 10.1007/BF01309867. [DOI] [PubMed] [Google Scholar]
- 24.Veits J, Weber S, Stech O, Breithaupt A, Gräber M, Gohrbandt S, Bogs J, Hundt J, Teifke JP, Mettenleiter TC, Stech J. 2012. Avian influenza virus hemagglutinins H2, H4, H8, and H14 support a highly pathogenic phenotype. Proc Natl Acad Sci USA 109:2579–2584. 10.1073/pnas.1109397109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerlach T, Hensen L, Matrosovich T, Bergmann J, Winkler M, et al. 2017. pH optimum of hemagglutinin-mediated membrane fusion determines sensitivity of influenza A viruses to the interferon-induced antiviral state and IFITMs. J Virol 91:e00246-17. 10.1128/JVI.00246-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X, Shi Y, Gao F, Xiao H, Wang M, Qi J, Gao GF. 2012. Insights into avian influenza virus pathogenicity: the hemagglutinin precursor HA0 of subtype H16 has an alpha-helix structure in its cleavage site with inefficient HA1/HA2 cleavage. J Virol 86:12861–12870. 10.1128/JVI.01606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harbig A, Mernberger M, Bittel L, Pleschka S, Schughart K, Steinmetzer T, Stiewe T, Nist A, Böttcher-Friebertshäuser E. 2020. Transcriptome profiling and protease inhibition experiments identify proteases that activate H3N2 influenza A and influenza B viruses in murine airways. J Biol Chem 295:11388–11407. 10.1074/jbc.RA120.012635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Böttcher-Friebertshäuser E, Stein DA, Klenk H-D, Garten W. 2011. Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide-conjugated morpholino oligomer targeting the hemagglutinin-activating protease TMPRSS2. J Virol 85:1554–1562. 10.1128/JVI.01294-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baron J, Tarnow C, Mayoli-Nüssle D, Schilling E, Meyer D, Hammami M, Schwalm F, Steinmetzer T, Guan Y, Garten W, Klenk H-D, Böttcher-Friebertshäuser E. 2013. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J Virol 87:1811–1820. 10.1128/JVI.02320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laporte M, Stevaert A, Raeymaekers V, Boogaerts T, Nehlmeier I, et al. 2019. Hemagglutinin cleavability, acid stability, and temperature dependence optimize influenza B virus for replication in human airways. J Virol 94:e01430-19. 10.1128/JVI.01430-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galloway SE, Reed ML, Russell CJ, Steinhauer DA. 2013. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog 9:e1003151. 10.1371/journal.ppat.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409–417. 10.1016/S0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 33.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1870. 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 34.Gambaryan AS, Matrosovich TY, Boravleva EY, Lomakina NF, Yamnikova SS, Tuzikov AB, Pazynina GV, Bovin NV, Fouchier RAM, Klenk H-D, Matrosovich MN. 2018. Receptor-binding properties of influenza viruses isolated from gulls. Virology 522:37–45. 10.1016/j.virol.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Kawaoka Y, Naeve CW, Webster RG. 1984. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology 139:303–316. 10.1016/0042-6822(84)90376-3. [DOI] [PubMed] [Google Scholar]
- 36.Lambertz RLO, Pippel J, Gerhauser I, Kollmus H, Anhlan D, Hrincius ER, Krausze J, Kühn N, Schughart K. 2018. Exchange of amino acids in the H1-haemagglutinin to H3 residues is required for efficient influenza A virus replication and pathology in Tmprss2 knock-out mice. J Gen Virol 99:1187–1198. 10.1099/jgv.0.001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wille M, Robertson GJ, Whitney H, Bishop MA, Runstadler JA, Lang AS. 2011. Extensive geographic mosaicism in avian influenza viruses from gulls in the northern hemisphere. PLoS One 6:e20664. 10.1371/journal.pone.0020664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhagen JH, Eriksson P, Leijten L, Blixt O, Olsen B, et al. 2021. Host range of influenza A virus H1 to H16 in Eurasian ducks based on tissue and receptor binding studies. J Virol 95:e01873-20. 10.1128/JVI.01873-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.List K, Szabo R, Molinolo A, Nielsen BS, Bugge TH. 2006. Delineation of matriptase protein expression by enzymatic gene trapping suggests diverging roles in barrier function, hair formation, and squamous cell carcinogenesis. Am J Pathol 168:1513–1525. 10.2353/ajpath.2006.051071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberst MD, Singh B, Ozdemirli M, Dickson RB, Johnson MD, Lin C-Y. 2003. Characterization of matriptase expression in normal human tissues. J Histochem Cytochem 51:1017–1025. 10.1177/002215540305100805. [DOI] [PubMed] [Google Scholar]
- 41.Peacock THP, James J, Sealy JE, Iqbal M. 2019. A global perspective on H9N2 avian influenza virus. Viruses 11:620. 10.3390/v11070620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ko C-J, Huang C-C, Lin H-Y, Juan C-P, Lan S-W, Shyu H-Y, Wu S-R, Hsiao P-W, Huang H-P, Shun C-T, Lee M-S. 2015. Androgen-induced TMPRSS2 activates matriptase and promotes extracellular matrix degradation, prostate cancer cell invasion, tumor growth, and metastasis. Cancer Res 75:2949–2960. 10.1158/0008-5472.CAN-14-3297. [DOI] [PubMed] [Google Scholar]
- 43.Beaulieu A, Gravel É, Cloutier A, Marois I, Colombo É, Désilets A, Verreault C, Leduc R, Marsault É, Richter MV. 2013. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J Virol 87:4237–4251. 10.1128/JVI.03005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamilton BS, Gludish DWJ, Whittaker GR. 2012. Cleavage activation of the human-adapted influenza virus subtypes by matriptase reveals both subtype and strain specificities. J Virol 86:10579–10586. 10.1128/JVI.00306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puente XS, Sánchez LM, Overall CM, López-Otín C. 2003. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet 4:544–558. 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 46.Magnen M, Elsässer BM, Zbodakova O, Kasparek P, Gueugnon F, Petit-Courty A, Sedlacek R, Goettig P, Courty Y. 2018. Kallikrein-related peptidase 5 and seasonal influenza viruses, limitations of the experimental models for activating proteases. Biol Chem 399:1053–1064. 10.1515/hsz-2017-0340. [DOI] [PubMed] [Google Scholar]
- 47.Dittmann M, Hoffmann H-H, Scull MA, Gilmore RH, Bell KL, Ciancanelli M, Wilson SJ, Crotta S, Yu Y, Flatley B, Xiao JW, Casanova J-L, Wack A, Bieniasz PD, Rice CM. 2015. A serpin shapes the extracellular environment to prevent influenza A virus maturation. Cell 160:631–643. 10.1016/j.cell.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Straus MR, Kinder JT, Segall M, Dutch RE, Whittaker GR. 2020. SPINT2 inhibits proteases involved in activation of both influenza viruses and metapneumoviruses. Virology 543:43–53. 10.1016/j.virol.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamilton BS, Chung C, Cyphers SY, Rinaldi VD, Marcano VC, Whittaker GR. 2014. Inhibition of influenza virus infection and hemagglutinin cleavage by the protease inhibitor HAI-2. Biochem Biophys Res Commun 450:1070–1075. 10.1016/j.bbrc.2014.06.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quesada V, Velasco G, Puente XS, Warren WC, López-Otín C. 2010. Comparative genomic analysis of the zebra finch degradome provides new insights into evolution of proteases in birds and mammals. BMC Genomics 11:220. 10.1186/1471-2164-11-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, Lucas JM, Nelson PS, Pöhlmann S, Soilleux EJ. 2012. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One 7:e35876. 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long JS, Mistry B, Haslam SM, Barclay WS. 2019. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17:67–81. 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- 53.Peitsch C, Klenk H-D, Garten W, Böttcher-Friebertshäuser E. 2014. Activation of influenza A viruses by host proteases from swine airway epithelium. J Virol 88:282–291. 10.1128/JVI.01635-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zmora P, Molau-Blazejewska P, Bertram S, Walendy-Gnirß K, Nehlmeier I, Hartleib A, Moldenhauer A-S, Konzok S, Dehmel S, Sewald K, Brinkmann C, Curths C, Knauf S, Gruber J, Mätz-Rensing K, Dahlmann F, Braun A, Pöhlmann S. 2017. Non-human primate orthologues of TMPRSS2 cleave and activate the influenza virus hemagglutinin. PLoS One 12:e0176597. 10.1371/journal.pone.0176597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ovcharenko AV, Zhirnov OP. 1994. Aprotinin aerosol treatment of influenza and paramyxovirus bronchopneumonia of mice. Antiviral Res 23:107–118. 10.1016/0166-3542(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 56.Zhirnov OP, Klenk HD, Wright PF. 2011. Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral Res 92:27–36. 10.1016/j.antiviral.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 57.Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. 2012. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol 86:6537–6545. 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto M, Matsuyama S, Li X, Takeda M, Kawaguchi Y, Inoue J-I, Matsuda Z. 2016. Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother 60:6532–6539. 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bestle D, Heindl MR, Limburg H, van Lam van T, Pilgram O, et al. 2020. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance 3:e202000786. 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Böttcher E, Freuer C, Steinmetzer T, Klenk H-D, Garten W. 2009. MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine 27:6324–6329. 10.1016/j.vaccine.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 61.Steinmetzer T, Hardes K. 2018. The antiviral potential of host protease inhibitors, p 279–325. In Böttcher-Friebertshäuser E, Garten W, Klenk HD (ed), Activation of viruses by host proteases. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 62.Moulton HM, Moulton JD. 2010. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim Biophys Acta 1798:2296–2303. 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 63.Böttcher-Friebertshäuser E. 2018. Membrane-anchored serine proteases: host cell factors in proteolytic activation of viral glycoproteins, p 153–203. In Böttcher-Friebertshäuser E, Garten W, Klenk HD (ed), Activation of viruses by host proteases. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 64.Matrosovich M, Matrosovich T, Uhlendorff J, Garten W, Klenk H-D. 2007. Avian-virus-like receptor specificity of the hemagglutinin impedes influenza virus replication in cultures of human airway epithelium. Virology 361:384–390. 10.1016/j.virol.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 65.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA 97:6108–6113. 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abes S, Moulton HM, Clair P, Prevot P, Youngblood DS, Wu RP, Iversen PL, Lebleu B. 2006. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J Control Release 116:304–313. 10.1016/j.jconrel.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 67.Kim TS, Heinlein C, Hackman RC, Nelson PS. 2006. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol Cell Biol 26:965–975. 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beyer HM, Gonschorek P, Samodelov SL, Meier M, Weber W, Zurbriggen MD. 2015. AQUA cloning: a versatile and simple enzyme-free cloning approach. PLoS One 10:e0137652. 10.1371/journal.pone.0137652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1<br>. Download JVI.00906-21-s0001.pdf, PDF file, 1.7 MB (1.7MB, pdf)