FIG 1.
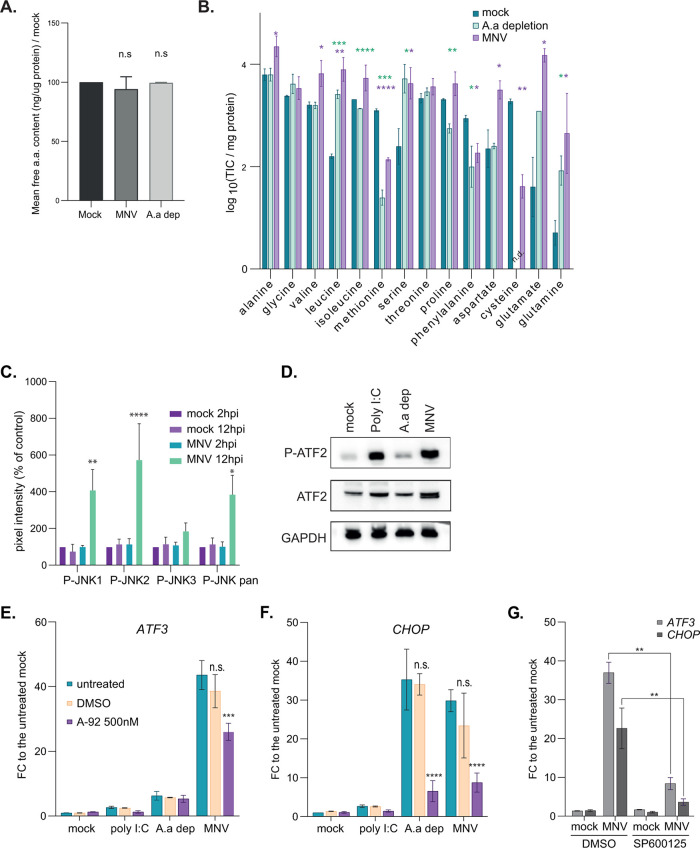
MNV infection is associated with an amino acid (a.a.) depletion phenotype. (A and B) Quantification of free amino acids in RAW264.7 cells mock (dark green) and MNV infected (magenta) for 10 h or incubated in GCM-depleted medium (light green). (A) There were no significant differences between the total concentration of free amino acids quantified using a colorimetric assay. By contrast GC-MS (B) measured significant differences in the pool sizes of individual amino acids (TIC, total ion current). Values are means ± standard errors of the mean (SEM) (n = 3) submitted to t test pairwise comparison. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.1. The mass-to-charge ratios for analyzed amino acids are alanine 260, glycine 246, valine 288, norvaline 288, leucine 274, isoleucine 274, methionine 320, serine 390, threonine 404, proline 258, phenylalanine 336, aspartate 418, cysteine 406, glutamate 432, ornithine 417, asparagine 417, lysine 431, glutamine 431, arginine 442, histidine 440, tyrosine 466, tryptophan 375; n.d., not determined. (C) Stress related JNK1/2 are activated during MNV replication in RAW264.7 cells. Bar plot (n = 3) of the mean ± standard deviation (SD) of the results of phosphoantibody array assay (n = 3) at 2 h p.i. (light) and 12 h p.i. (dark) in mock and cells infected with MNV at an MOI of 10 (green and purple bars, respectively). Statistical analysis results shown above the plots. *, P < 0.1; **, P < 0.01; ****, P < 0.0001. (D) Western blot analysis of the activation of ATF2 in mock cells, cells treated with 20 μg/ml of poly(I·C), amino acid-starved cells and MNV-infected cells at 10 h p.i. (MOI, 10). (E and F) GCN2 is partially responsible for the genetic reprogramming in MNV-infected cells. Bar plots (n = 4) of the mean ± SEM of transcript upregulation analysis by qRT-PCR for ATF3 mRNA (E) and CHOP mRNA (F). Experiment performed on total transcripts from RAW264.7 cells treated as in panel B for 10 h with or without 500 nM GCN2 inhibitor A92. Purified RNAs were reverse transcribed using poly(dT) primer and quantitative PCR (qPCR) performed using exon-junction spanning pair of PCR primer. Statistical significance between untreated and treated samples calculated using analysis of variance (ANOVA) two-way multiple comparison tool in GraphPad shown above the bars. ****, P < 0.0001; ***, P < 0.001; n.s., not significant. (G) Bar plots (n = 3) of the mean ± SEM of transcript upregulation analysis by qRT-PCR for ATF3 mRNA and CHOP mRNAs following JNK inhibitions. Experiment performed on total transcripts from RAW264.7 cells treated as in panel E for 10 h with or without 20 μM JNK inhibitor SP600125. Statistical significance between untreated and treated samples calculated using ANOVA two-way multiple comparison tool in GraphPad shown above the bars. ****, P < 0.0001; ***, P < 0.001; n.s., not significant.