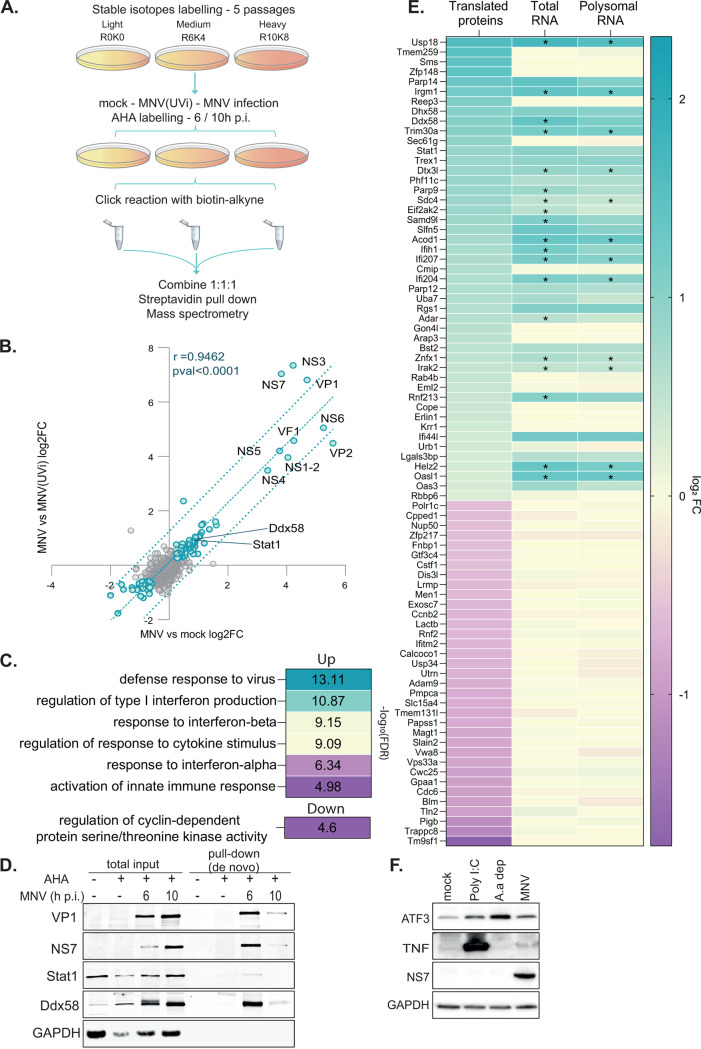
FIG 6.
Validation of the results of the translatomic data analysis by mass spectrometry analysis of the nascent proteome in RAW264.7 cells. Genome-wide analysis of the nascent proteome by biorthogonal labeling of the nascent protein and mass spectrometry allowing an extensive validation of the active translatome identified by RNA sequencing. (A) Diagram of the experimental design of the biorthogonal dual labeling proteomics assay as described in reference 53 from SILAC labeling to mass spectrometry. (B) Scatterplot of the results of the differential expression analysis in the contrasts MNV-/mock-infected cells (x axis) versus MNV-/MNV(UVi)-infected cells (y axis) at 10 h p.i. showing the enrichment for viral structural and nonstructural proteins as well as a subset of MNV-induced differentially expressed host proteins with similar behavior in both contrasts as shown by the correlation analysis. The differential expression analysis was performed on all samples at 10 h p.i. using a pairwise ratio, and results expressed as log2(FC) of the complete gene data set were used for the biorthogonal projection, significantly regulated protein (green dots) and not significantly regulated protein (gray dots). Correlation analysis using the Pearson method showed a high degree of correlation between the two contrasts (R2 > 0.89; pval < 0.0001). Dotted blue line, linear regression and IC95 of significance. (C) Clustered results of the gene ontology analysis showing enrichment of the upregulated host proteins in antiviral innate immune response terms and enrichment of the downregulated host proteins in regulation of the cell cycle. The gene annotation analysis was performed in Biological Process and KEGG pathway terms using the ClueGO plug-in on the Cytoscape platform for all genes significantly regulated at 10 h p.i. and ordered by decreasing corresponding −log10(FDR). (D) Validation of the proteomics analysis setup and results. Representative Western blot against viral proteins and two host proteins upregulated in the proteomics at 10 h p.i. on the inputs (total input) and pulldown fractions of AHA-labeled and lysed RAW264.7 cells at 6 and 10 h p.i. followed by click reaction and streptavidin pulldown, using AHA-unlabeled cells as a negative control. (E) Comparison of the proteomic and translatomic results in RAW264.7, showing both differential expression and differential protein stability in MNV-infected cells at 10 h p.i., with a decrease of stability for all of the proteins identified as downregulated in the proteomic analysis. Heatmap of comparison of the log2(FC) of the genes identified by proteomic in MNV versus MNV(UVi) (translated proteins) against their corresponding log2(FC) in total (total RNA) and polysomal fraction (polysomal RNA) generated from the translatomic data. Stars represent significant differential expression as described in Fig. 1A. (F) Validation by Western blotting of some of the upregulated genes in MNV-infected RAW264.7 cells identified by RNA sequencing but absent from the proteomic data set. Mock and MNV-infected cells lysates were run on SDS-Page alongside poly(I·C)-treated cells at 20 μg/ml and amino acid-starved cells for 10 h as positive control for the upregulation of TNF and ATF3 respectively.