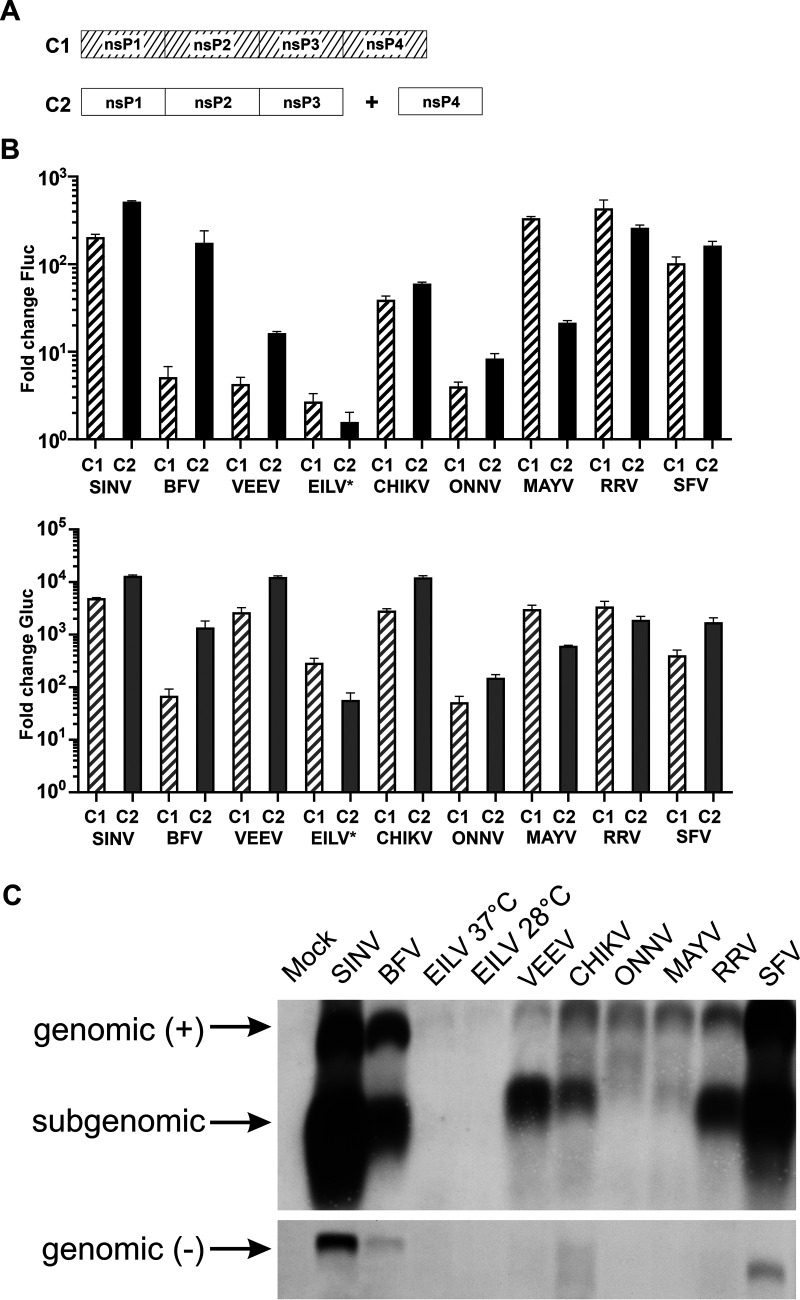
FIG 2.
Single- and two-component replicases of alphaviruses have similar activities. (A). Schematic presentation of expressed replicase precursors. C1, single-component replicase from plasmids CMV-P1234-SINV and so on; C2, two-component replicase from plasmids CMV-P123-SINV+ CMV-ubi-nsP4-SINV and so on. (B) HEK293T cells in 96-well plates were cotransfected with matching pairs of CMV-P1234 and HSPolI-FG plasmids (C1) or with matching combinations of CMV-P123, CMV-ubi-nsP4 (in a 1:1 molar ratio), and HSPolI-FG plasmids (C2). As the negative control, CMV-P1234GAA, which encodes polyprotein lacking RNA polymerase activity, was used instead of CMV-P1234. Cells were incubated at 37°C and lysed for 18 h p.t.; cells transfected with plasmids containing sequences from EILV were incubated at 28°C and lysed for 48 h p.t. Fluc (marker of replication) and Gluc (marker of transcription) activities produced by active replicases were normalized to the P1234GAA controls. The value obtained for the P1234GAA controls was taken as 1. The means ± standard deviation (SD) of three independent experiments are shown. (C) HEK293T cells in 12-well plates were cotransfected with matching combinations of CMV-P123, CMV-ubi-nsP4 (in 1:1 molar ratio), and HSPolI-FG plasmids; control cells were mock-transfected. Cells were incubated as described for panel B, after which total RNA was extracted and analyzed by Northern blotting. Full-length “genomic” template RNA of positive (+) and negative (–) polarity and subgenomic RNA are indicated. Note that transcripts made by human RNA polymerase I using HSPolI-FG plasmids as templates comigrate with replicase-generated positive-strand genomic RNA and are detected by the same probe. The experiment was repeated twice with similar results; data from one experiment is shown.