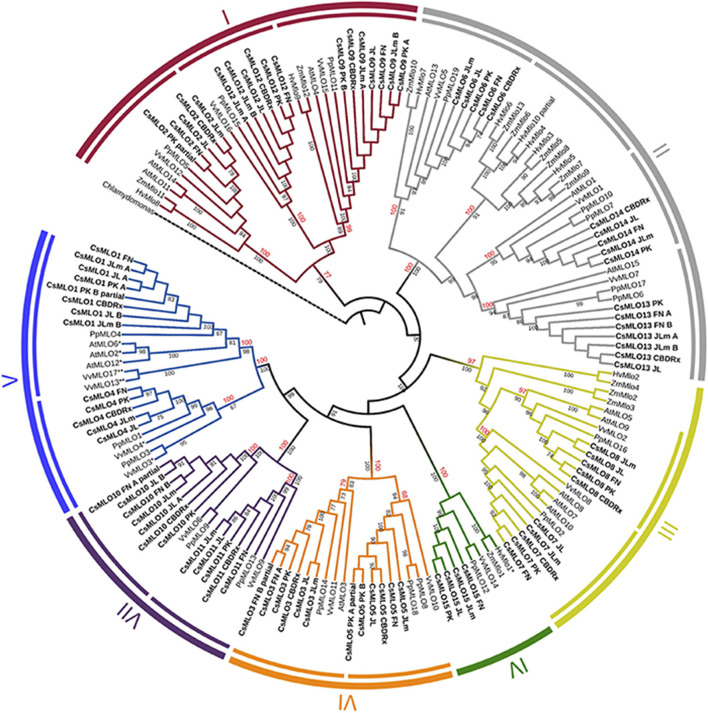
FIGURE 3.
Phylogenetic relationships of CsMLOs based on maximum likelihood analysis. Phylogenetic tree of manually curated CsMLO proteins (bold) with MLO proteins from selected species (Arabidopsis thaliana, Prunus persica, Vitis vinifera, Hordeum vulgare, and Zea mays). Chlamydomonas reinhardtii was used as an outgroup. Phylogenetic relationships were estimated using the maximum likelihood method implemented in PhyML + SMS with 1,000 bootstrap independent replicates. The seven defined clades are indicated, as well as potential subclades identified in this study (inner circles). Number on a node indicates the percentage of bootstrap when higher than 65% (black), or the posterior probabilities of major clades and subclades, according to a Bayesian phylogenetic inference performed on the same alignment (red) (Supplementary Figure 1). MLOs with one asterisk (*) have been experimentally demonstrated to be required for PM susceptibility (Büschges et al., 1997; Feechan et al., 2008; Wan et al., 2020), while MLOs with two asterisks (**) have been identified as main probable candidates for PM susceptibility (Pessina et al., 2016).