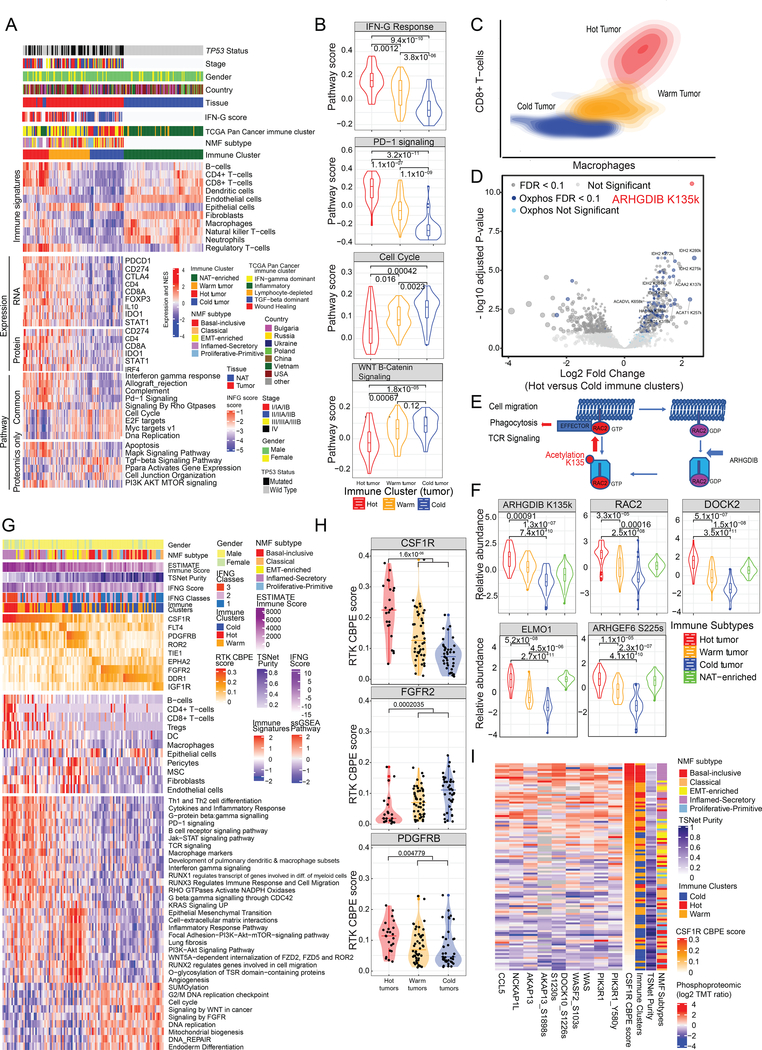
Figure 6: Immune Landscape of LSCC.
A. Heatmaps illustrate cell type compositions and activities of selected individual genes/proteins and pathways across the four immune clusters: Hot, Warm, and Cold tumor and NAT-enriched. Successive heatmaps illustrate xCell immune signatures, mRNA and protein expression of key immune-related markers, and ssGSEA pathway scores based on global proteomic data for biological pathways that were differentially regulated in immune groups based on both mRNA and global protein abundance (Common) or on global protein abundance alone (Proteomics only).
B. Pathway scores of key pathways differentially expressed across the immune clusters. Wilcoxon p-values for the individual comparisons are provided on top.
C. Contour plot of two-dimensional density based on Macrophage (x-axis) and CD8 T-cell scores (y-axis) showing the variation in these cell types’ distributions observed across the different immune clusters.
D. Acetylsites differentially expressed between Hot and Cold tumors. Acetylsites of genes contained in the Hallmark Oxidative Phosphorylation pathway are highlighted in blue, ARHGDIB K135 is highlighted in red, and remaining sites are in gray. Darker color designates significant sites (FDR < 0.1).
E. Regulation of Rho GTPase signaling including K135 acetylation of ARHGDIB.
F. Global protein abundance of RAC2, DOCK2 and ELMO1, acetylproteome abundance of ARHGDIB K135k and phosphorylation abundance of ARHGEF6 at Serine 225 in immune clusters. Wilcoxon p-values are reported.
G. RTK CBPE scores for 108 tumor samples and associated xCell signatures and pathway scores. The first heatmap shows CBPE scores of key RTKs, the second xCell signatures (Aran et al., 2017) and the third pathway scores based on global protein abundance.
H. Distribution of RTK CBPE scores for CSF1R, PDGFRB and FGFR2 stratified by immune clusters. Significance values (two-sided Wilcoxon test) between Hot clusters and combined Warm and Cold clusters are indicated on the violin plots.
I. Heatmap showing proteins and phosphosites correlated with CSF1R CBPE scores that are known to be involved in immune evasion. See also Figure S6 and Table S6