Figure 3. MFZ 2–24 and DAT Induced Fit Docking complexes and molecular dynamics simulation.
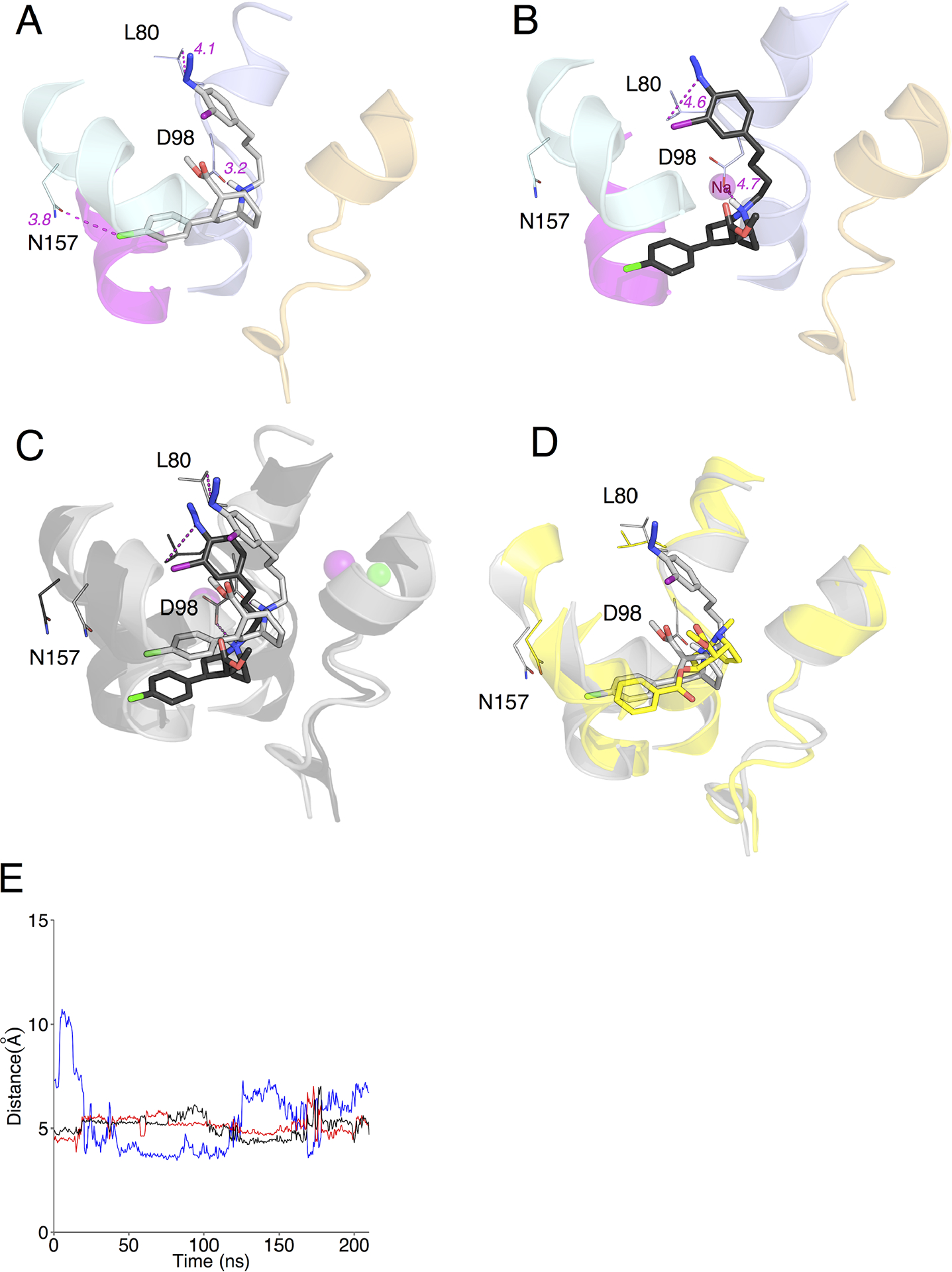
(A) Best IFD rDAT/MFZ 2–24-docked complex based on energy and constraint fulfillment. Image depicts the LeuT-based rDAT homology model from the crystal structure of the open-to-out template 3F3A. (B) Molecular dynamics simulation of the IFD rDAT/MFZ 2–24-docked complex. (C) Superimposition of IFD-docked MFZ 2–24 before (grey) and after (black) simulation. (D) Superimposition of IFD-docked MFZ 2–24 pre-simulation (grey) and cocaine (yellow) from Drosophila DAT co-crystal. (E) Time-dependent changes in distances following molecular dynamics simulation between the protonated nitrogen of MFZ 2–24 and the side chain oxygen atom of Asp79 (blue and black traces for ∂O1 and ∂O2) and the phenyl chloride of MFZ 2–24 and the amide nitrogen of Asn157 (red traces).
