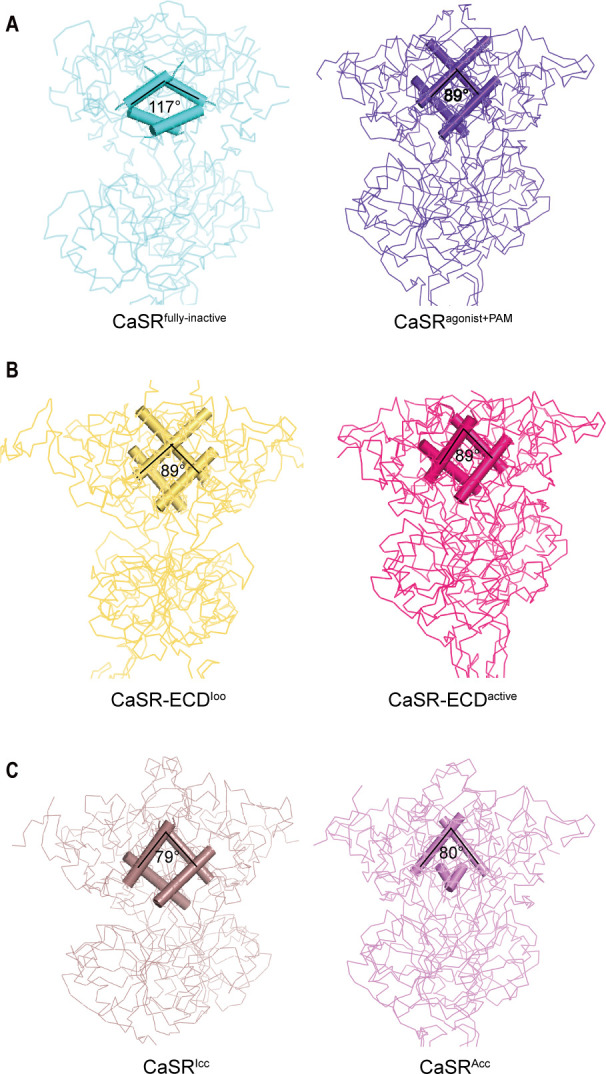
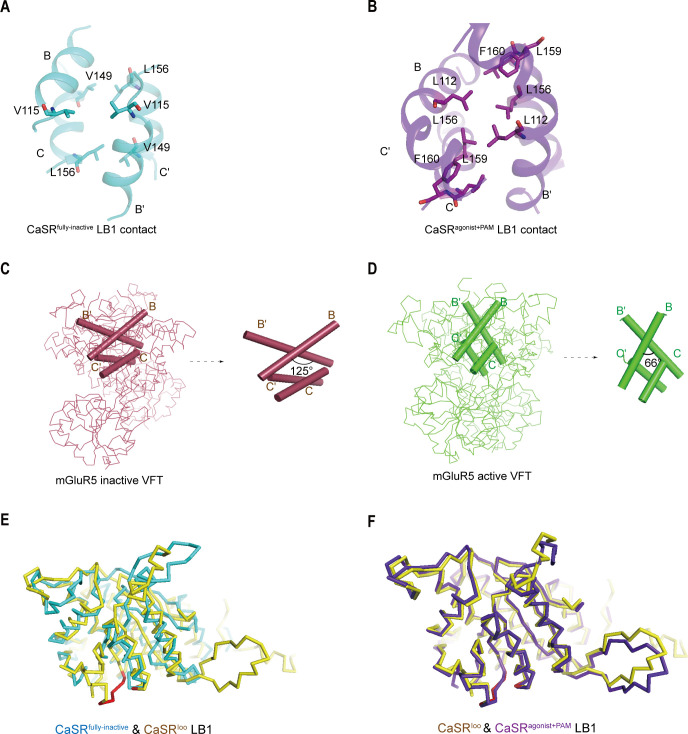
Figure 4. Comparisons of intersubunit LB1 domains interfaces in the inactive and active states of CaSR.
(A) Left panel: The Cα trace of VFT module of CaSRfully inactive cryo-EM structure (cyan). The B-C Helix angle is 117°. Right panel: The Cα trace of VFT module of CaSRagonist+PAM cryo-EM structure (purple). The B-Helix angle is 89°. (B) Left panel: The Cα trace of VFT module of crystal structure of CaSR-ECDIoo (yellow) (PDB:5K5T). The B-Helix angle is 89°. Right panel: The Cα trace of VFT module of CaSR-ECDactive crystal structure (red) (PDB:5K5S). The B-Helix angle is 89°. (C) Left panel: The Cα trace of VFT module of CaSRIcc cryo-EM structure (brown) (PDB:7DTW). The B-Helix angle is 79°. Right panel: The Cα trace of VFT module of CaSRAcc cryo-EM structure (lavender) (PDB:7DTV). The B-Helix angle is 80°.