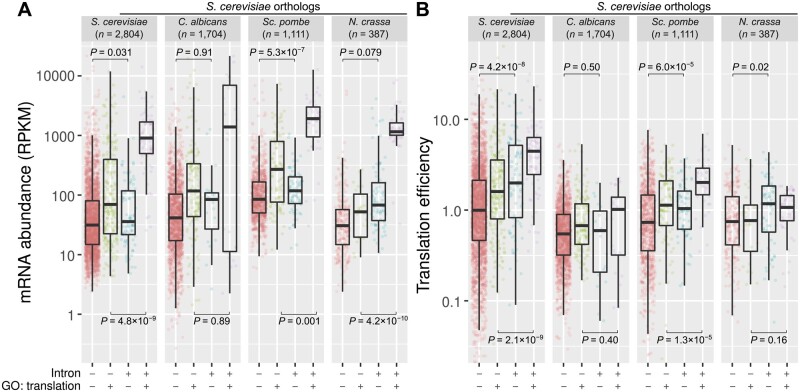
Fig. 9.
Intron-containing genes have higher levels of mRNA expression and translation efficiency. Matched RNA-seq and ribosome profiling data sets were used in this analysis (supplementary Table S7, Supplementary Material online). (A) mRNA abundance was calculated using RPKM. (B) Translation efficiency was determined by the ratio of ribosome-protected fragments and RNA-seq read counts that were normalized by the respective library sizes. Saccharomyces cerevisiae orthologs were grouped into four classes: (++) intron-containing genes annotated with the GO term “translation,” (+−) intron-containing genes annotated with other GO terms, (−+) intronless genes annotated with the GO term “translation,” and (−−) intronless genes annotated with other GO terms. The levels of mRNA expression and translation efficiency between intron-containing and intronless genes were compared using Welch two-sample t-test. C. albicans, Candida albicans; N. crassa, Neurospora crassa; GO, gene ontology; RPKM, reads per kilobase per million mapped reads; S. cerevisiae, Saccharomyces cerevisiae; Sc. pombe, Schizosaccharomyces pombe.