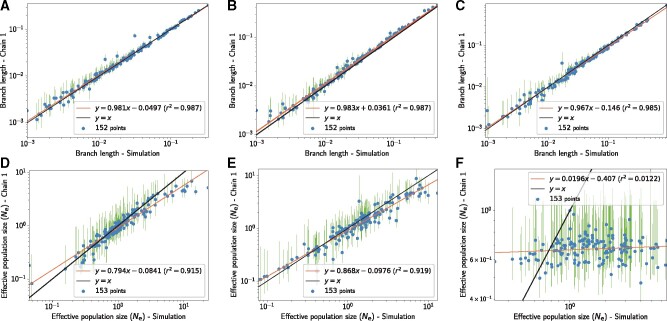
Fig. 2.
(A–C) Branch lengths in expected number of substitutions per site. (D–F) values across nodes (including the leaves) relative to at the root. From left to right: Simulation under the mutation–selection approximation (A, D), under a Wright–Fisher model accounting for small population size effects (5,000 individuals at the root), site linkage and short term fluctuation of (B, E), and finally accounting for site epistasis in the context of selection for protein stability (C, F). The tree root is 150 My old, where the initial population starts with a mutation rate of per site per generation and generation time of 10 years. These experiments confirm that signal in the placental mammalian tree can allow to reliably infer the direction of change in , even if linkage disequilibrium, short term fluctuation of and finite population size effects are not accounted for in the inference framework. However, the presence of epistasis between sites is a serious threat to the inference of .