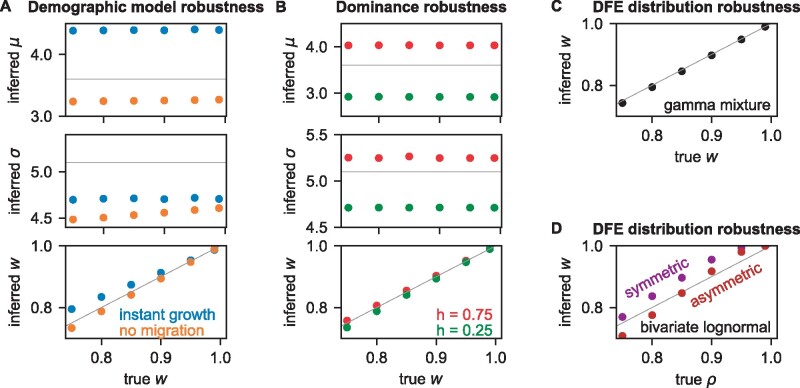
Fig. 2.
Robustness of joint DFE inference to model misspecification. Simulated neutral and selected data were generated under a demographic model with exponential growth and migration (supplementary table S1, Supplementary Material online), and lognormal mixture DFE models were fit to the data. The DFE parameters are: μ, the mean log population-scaled selection coefficient; σ, the standard deviation of those log coefficients; and w, the correlation of the DFE. The gray lines indicate true values, and the data plotted in these figures can be found in supplementary tables S4–S6, Supplementary Material online. (A) In this case, simpler demographic models with instantaneous growth or symmetric migration were fit to the neutral data. The resulting misspecified model was then used when inferring the DFE. This misspecification biased μ and σ, but not w. (B) In this case, selected data were simulated assuming dominant or recessive mutations, but the DFE was inferred assuming no dominance (h = 0.5). Again, μ and σ are biased, but w is not. (C) In this case, selected data were simulated using a mixture of gamma distributions. When these data were fit using our mixture of lognormal distributions, w was not biased. (D) In this case, selected data were simulated using bivariate lognormal models, with either symmetric or asymmetric marginal distributions. When these data were fit using our symmetric mixture of lognormal distributions, w was only slightly biased.