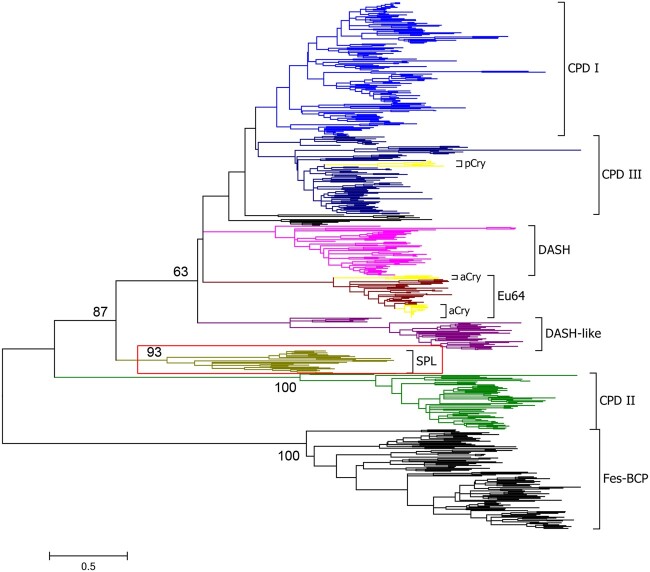
Fig. 1.
Phylogenetic tree of the cryptochrome/photolyase family (CPF). The evolutionary history was inferred by using the maximum likelihood method based on the Le_Gascuel_2008 with Freq. (+F) model (Le and Gascuel 2008) and 500 bootstrap iterations. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter=1.3083]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 2.0619% sites). Numbers at nodes represent the bootstrap support percentage values. The CPF proteins were divided into eight main groups: class I CPD photolyases (CPD I), class III CPD photolyases (CPD III), DASH proteins (DASH), eukaryotic 6-4 photolyases (Eu64), DASH-like proteins (DASH-like), short photolyase-likes (SPL), class II CPD photolyases (CPD II), and FeS-BCP proteins (FeS-BCP). Plant cryptochromes (pCry) and animal cryptochromes (aCry) were clustered as the subgroups of CPD III and Eu6-4, respectively. The SPL group was highlighted with a red box, which was identified in this report.