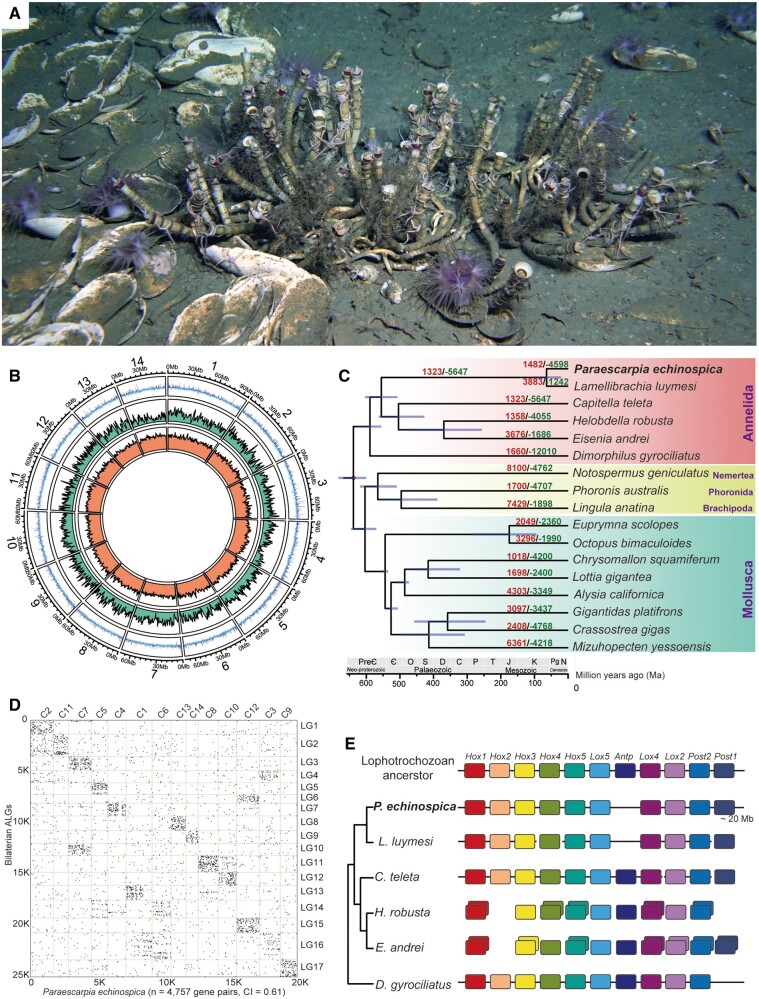
Fig. 1.
Natural habitat, genomic structure, and phylogenetic position of Paraescarpia echinospica. (A) Photograph showing a colony of P. echinospica individuals in the field (each tube roughly 30 cm in length). Photos taken by Haima ROV crew. (B) Circus plot of 14 pseudochromosomal linkage groups showing marker distribution at 1 Mb sliding windows from outer to inner circle: GC content, gene density, and repeat density. (C) Phylogenetic tree showing the phylogenetic position of P. echinospica in Lophotrochozoa. The tree was constructed using a maximum likelihood method with LG + I + G model and calibrated with fossil records at four nodes shown with a red dot. The purple lines on the nodes indicate divergence time with a 95% confidence interval. The numbers on each branch indicate gene family expansion (red) and contraction (green). C, Carboniferous; Є, Cambrian; D, Devonian; J, Jurassic; K, Cretaceous; M, Mesozoic; N, Neogene; O, Ordovician; P, Permian; Pg, Paleogene; PreЄ, Precambrian; S, Silurian; T, Triassic. (D) Macrosynteny comparison dot plots between the P. echinospica genome and the 17 presumed bilaterian ancient linkage groups (ALGs), suggesting that the chromosomes of P. echinospica have a conserved macrosynteny compared with the most recent common ancestor of Lophotrochozoa. Each dot represents the mutual protein best match between the two species determined by BlastP. (E) Schematic representation of the Hox cluster organization in annelids, with the putative ancestral Lophotrochozoa Hox cluster on the top. Each Hox orthologous group is colored differently.