Fig. 5 |. The Dcp2 C-terminus stabilizes an autoinhibited conformation required for regulation of decapping in condensates.
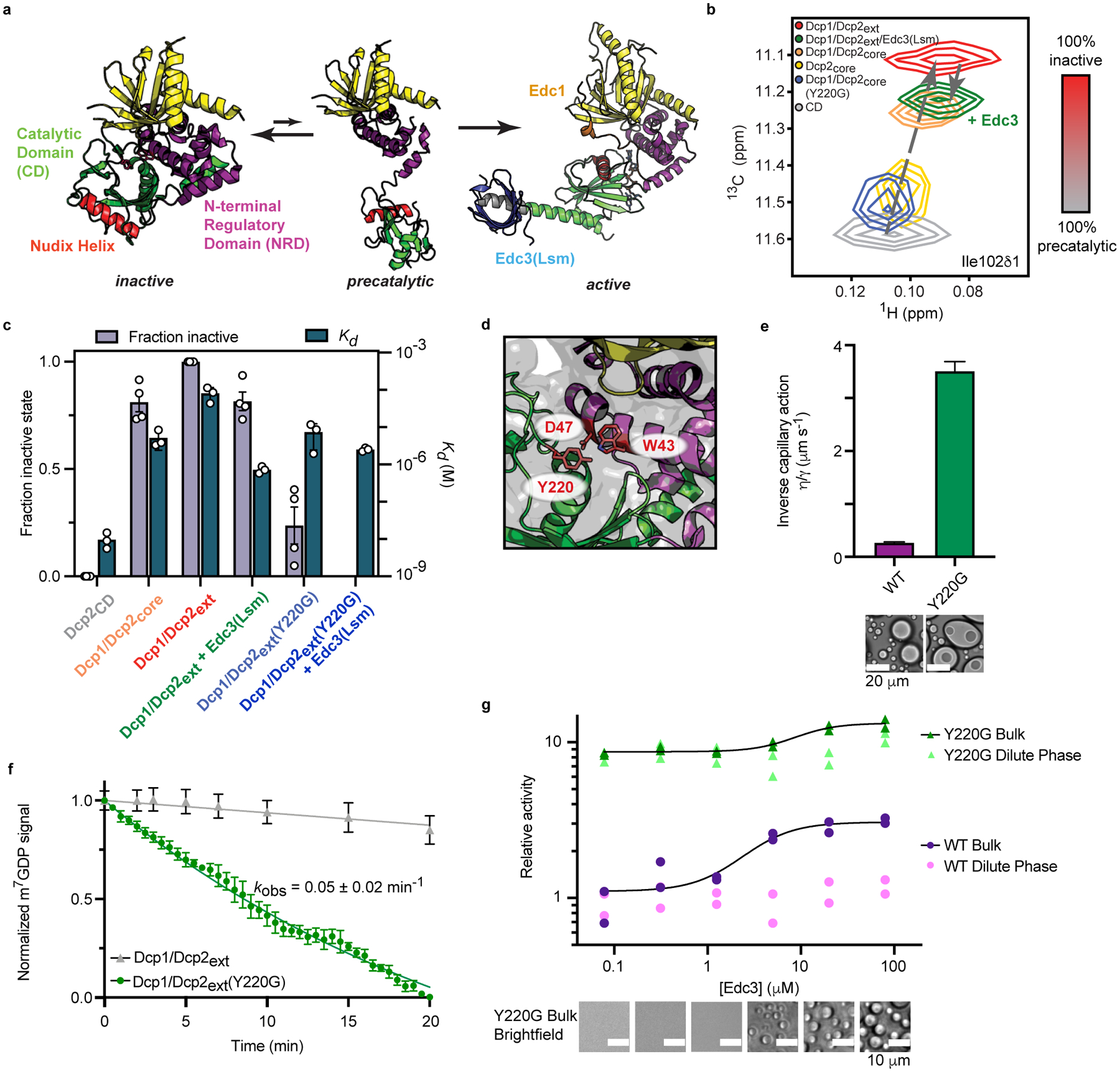
a, Dcp2 conformation is in a fast equilibrium between inactive and precatalytic states (PDB: 2QKM). Activators Edc1 and Edc3 stabilize the active state (PDB: 6AM0). Dcp1/Dcp2core is colored as in Figure 1a and the Nudix Helix, which contains catalytic residues is colored red. b, 1H/13C-HSQC of methyl group in Ile102 undergoes linear chemical shift changes toward the inactive state when the Dcp2 regulatory domain, Dcp1, and the Dcp2 C-terminus are added. Edc3 and Y220G mutation in the Dcp2 catalytic domain revert the chemical shift toward the precatalytic state. c, The population of the inactive state correlates with weakened RNA binding by Dcp1/Dcp2. Data from four NMR resonances and three independent RNA binding experiments are presented as mean ± s.e.m. d, Y220 residue in the Dcp2 catalytic domain occludes residues critical for m7G recognition (W43 and D47) in the inactive state. e, Dcp1/Dcp2ext(Y220G) droplets do not relax to a spherical state after fusion, contain subcompartments, and exhibit a ten-fold greater viscosity-to-surface tension ratio relative to wild-type Dcp1/Dcp2ext droplets. Protein concentration is 100 μM. Reported error is standard error of the fit to data in Extended Data Fig. 5c. f, Dcp1/Dcp2ext(Y220G) increases decapping of dual-labeled RNA substrate 10-fold in droplets. Wild-type Dcp1/Dcp2ext data from Fig. 3g is reproduced for comparison. m7GDP intensity is normalized to initial fluorescence signal. Data are presented as mean ± s.e.m for twenty droplets examined over two independent experiments. g, The Y220G mutation activates decapping and minimizes contribution from decapping in condensates. Data presented are from two independent experiments and relative activity is determined by ratio of observed rates to wild-type Dcp1/Dcp2ext in absence of Edc3. Representative micrographs are from three independent experiments with similar results.
