Extended Data Fig. 1. Synthesis and decapping of dually labelled 5’ capped 35 nt RNA probe.
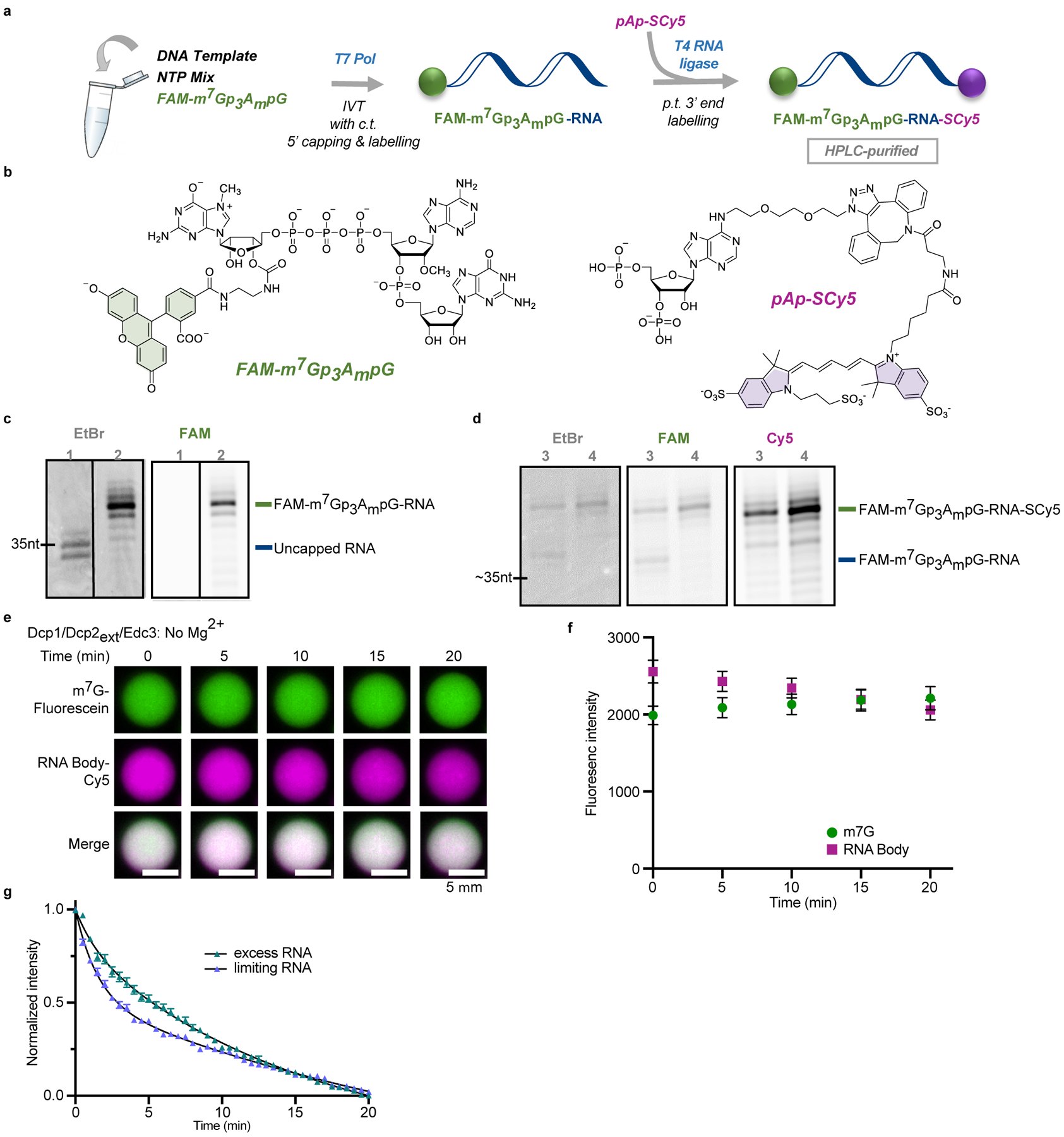
a, Overview of the labelling procedure, IVT – in vitro transcription, c.t. – co-transcriptional, p.t. – post-transcriptional; b, structures of the reagents used for the 5’ and 3’ end labelling; c, Analysis of purified 5’ capped & labelled RNA after IVT: lane 1 – reference uncapped RNA, lane 2 – RNA capped co-transcriptionally with fluorescent cap analog (FAM-m7Gp3AmpG). d, Labelling of the 3’ end of FAM-m7Gp3AmpG-RNA with pAp-SCy5 to yield dually labelled probe: lane 3 – crude dually labelled RNA after purification; lane 4 – HPLC-purified RNA probe. e, f, Co-localization of m7G cap (fluorescein) and RNA body (Cy5) in Dcp1/Dcp2ext/Edc3 condensates over twenty minutes demonstrates decapping does not occur in the absence of Mg2+, which is required for catalysis. g, Excess RNA slows initial rate of decapping two-fold in droplets formed with 1 μM Dcp1/Dcp2ext and 15 μM Edc3. Total RNA concentration is 100 nM when limiting and 20 μM when in excess. Representative micrographs and data in f, g are presented as mean ± s.e.m. for twenty droplets examined in two independent experiments with similar results. Error bars are not depicted when smaller than the data points.
